基于对虾东方病毒基因组序列的检测方法及其应用与流程
本发明属于水产养殖领域,具体涉及对虾东方病毒基因组序列的检测方法及其应用。
背景技术:
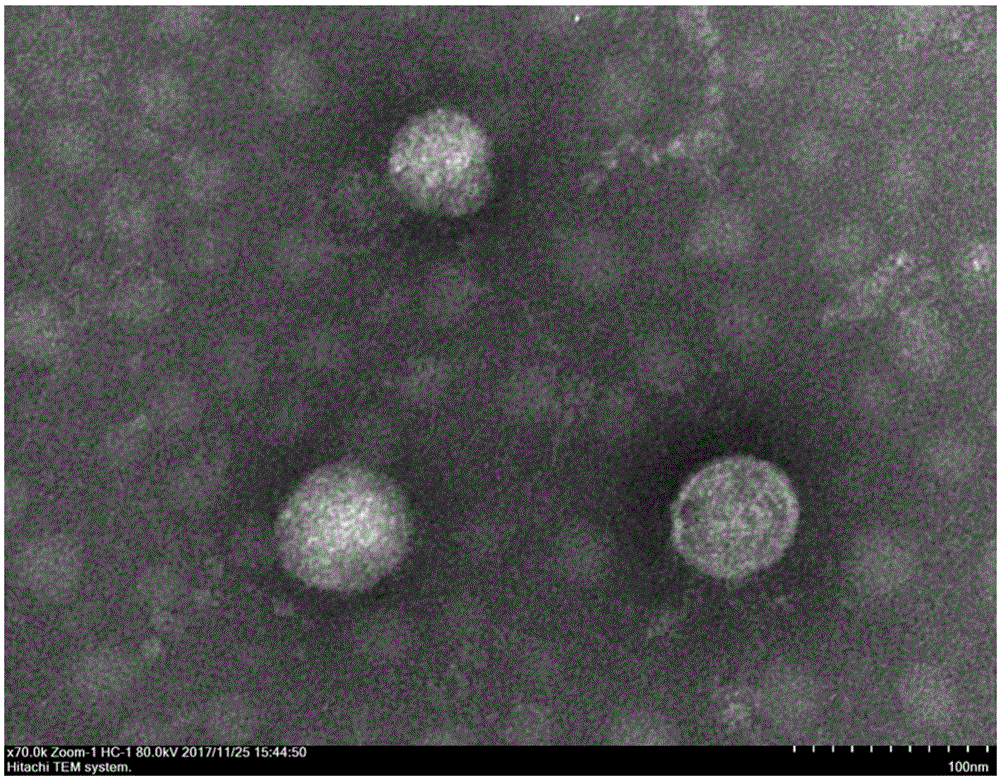
:国际病毒分类委员会第十次报告中新命名的布尼亚病毒目分为9科13属,科分别为费拉病毒科(feraviridae)、无花果花叶病毒科(fimoviridae)、汉坦病毒科(hantaviridae)、米卡多病毒科(jonviridae)、内罗病毒科(nairoviridae)、泛布尼亚病毒科(peribunyaviridae)、幻影病毒科(phasmaviridae)、白纤病毒科(phenuiviridae)和番茄斑萎病毒科(tospoviridae)。费拉病毒科仅有1个病毒属:正费拉病毒属(orthoferavirus),主要感染蚊子,不能感染脊椎动物细胞,属于昆虫宿主限制性病毒;无花果花叶病毒科包含伊马拉病毒属(emaravirus),可感染植物,并可通过植物嫁接或螨类等节肢动物传播;汉坦病毒科包含正汉坦病毒属(orthohantavirus),可经啮齿动物和食虫类动物进行传播,偶尔也感染人类;米卡多病毒科(jonviridae)包含正米卡多病毒属(orthojonvirus),能在多种蚊子细胞中复制扩增,但不能感染脊椎动物细胞,属于昆虫宿主限制性病毒;内罗病毒科包含正内罗病毒属(orthonairovirus),该科病毒大部分以蜱、蚊为媒介传播,部分病毒也可在虱子、蠓等节肢动物中传播,或者通过蜱虫在不同宿主之间进行传播。内罗病毒科在非洲、亚洲、澳洲、欧洲和美洲均有分布,传染性强、致死率高,能够引起多种严重的人类及动物传染性疾病;泛布尼亚病毒科由赫伯病毒属和正布尼亚病毒属(orthobunyavirus)组成,其中,正布尼亚病毒可感染人和反刍动物,可引起病人的如发热、脑炎和出血热等严重疾病;幻影病毒科包含正幻影病毒属(orthophasmavirus),仅感染蟑螂、尺蛾和蚊,属于昆虫宿主限制性病毒;白纤病毒科由格克病毒属、帕西病毒属(phasivirus)、白蛉病毒属(phlebovirus)和纤细病毒属(tenuivirus)组成,格克病毒属仅能感染蚊,尚未有感染其他脊椎动物和节肢动物的报道;帕西病毒属可感染苍蝇和蚊等节肢动物,尚未有感染脊椎动物的报道;白蛉病毒属病毒都是虫媒病毒,可通过白蛉、蜱、蠓和蚊子等节肢动物传播,感染多种哺乳动物和人类,严重影响人类健康;纤细病毒属是一类植物病毒,可在植物细胞内扩增,通过飞虱、叶蝉传播;番茄斑萎病毒科包含正番茄斑萎病毒属(orthotospovirus),可通过植物枝叶进行传播,可通过昆虫媒介蓟马在寄主植株间传播。1996年,澳大利亚学者在北昆士兰州毛利镇(mourilyan)的发病斑节对虾中首次鉴定了一株布尼亚病毒-毛利病毒(mourilyanvirus,mov),该病毒随后在东南亚及澳大利亚的日本对虾和斑节对虾中均有报道,已有研究证明毛利病毒对日本对虾有致病性。2018年澳大利亚学者sakuna报道了从红螯螯虾中发现了一株新的布尼亚病毒,系统发育进化树显示该病毒与泛布尼亚病毒科(peribunyaviridae)的正布尼亚病毒属(orthobunyavirus)的亲缘关系最近,并推测该病毒与红螯螯虾长途运输后的高死亡率相关。目前,发现能够感染甲壳纲动物的布尼亚病毒有限。技术实现要素:针对目前缺乏检测和防控甲壳类布尼亚病毒产品的问题,本发明提供一株对虾东方病毒(orientaviruspenaei),属于布尼亚病毒目的新种,能够感染甲壳类动物。本发明的另一目的在于提供一种检测上述新型布尼亚病毒株的试剂盒。为实现上述目的,本发明采用如下技术方案。一株对虾东方病毒(orientaviruspenaei),属于布尼亚病毒目,于2018年12月06日保藏于中国典型培养物保藏中心(cctcc),保藏地址为武汉大学保藏中心,保藏编号为cctccno.v201864。所述对虾东方病毒呈球形,由囊膜、衣壳和三节段的负链rna病毒组成,直径为70-120nm。能够感染中国对虾(fenneropenaeuschinensis)等,主要症状表现为活力降低,肝胰腺变浅。对虾东方病毒的基因组序列如seqidno:1、seqidno:2和seqidno:3所示;其中,l节段的序列如seqidno:1所示,m节段的序列如seqidno:2所示,s节段的序列如seqidno:3所示。一种检测类对虾东方病毒的试剂盒,所述类对虾东方病毒是指与对虾东方病毒同属的病毒。所述试剂盒是采用核酸扩增方法进行检测试剂盒或采用免疫反应进行检测的试剂盒。进一步的,所述试剂盒是采用核酸扩增技术进行检测试剂盒。由于东方病毒是一种单链rna病毒,基因组rna的合成直接由其依赖于rna的rna聚合酶进行复制,在rna复制过程中,依赖于rna的rna聚合酶缺乏3’-5’的纠错功能,导致基因组在自然复制时容易发生突变,基因组的整体或部分特异性序列的片段内可能存在≥70%的变异,因此在检测时,还需针对≥70%同源序列具有检测能力。上述试剂盒以(a)seqidno:1、seqidno:2或seqidno:3所示的rna序列或相应的dna序列全部或部分片段;或(b)(a)的互补序列;或(c)(a)或(b)的70%或以上同源的序列为特异性序列。上述特异性序列长度优选为12-3000nt;更优选的,长度为30nt-1200nt;最优选的,长度为40nt-600nt。所述试剂盒包含1对或1对以上的成对引物。所述试剂盒的引物可以是检测序列的特异性序列或含简并碱基的序列,还可以是包含锁核酸的序列。为了对≥70%同源的特异性序列进行检测时,最简单的方式是将存在单核苷酸多样性的突变位点视为相应的碱基的简并位点,从而可用包含多种突变碱基的简并碱基的序列作为引物序列;对于这种简并方式也可用次黄嘌呤或其他等效碱基代替。所述引物的长度为9nt-45nt;优选的,长度为15nt-30nt;更优选的,长度为18nt-25nt。进一步的,所述引物选自如seqidno:4-seqidno:26所示的任意2条或2条以上的正向和反向互补序列序列,或者2条之间的seqidno:1的特异性序列或其反向互补的特异性序列;或如seqidno:27-seqidno:34所示的任意2条或2条以上的正向和反向互补序列,或者2条之间的seqidno:2的特异性序列或其反向互补的特异性序列;或如seqidno:35-seqidno:40所示的任意2条或2条以上的正向和反向互补序列序列,或者2条之间的seqidno:3的特异性序列或其反向互补的特异性序列。当包含1对引物时,可采用常规pcr检测、sybrgreeni染色的实时荧光定量pcr检测、数字pcr检测或依赖于解旋酶的等温扩增(hda)检测。当包含1对以上引物时,可采用套式pcr或多重pcr检测,套式pcr采用2对嵌套的引物,其中外侧的1对引物作为套式pcr的外引物用于第一步扩增,内侧的1对引物作为套式pcr的内引物用于第二步扩增;多重pcr采用2对或更多对相互独立的引物,对多个位点或不同基因型的对虾东方病毒或其变异体进行检测。还可以按照环介导的等温扩增(lamp)引物的设计要求,以上述1对引物作为f3和b3引物,在其内侧,选择4条序列,通过组装设计成fip和bip引物,用于lamp检测;还可在单链环区在设计1对引物,作为增强扩增的lf和lb引物,用于更快速的lamp检测。在引物设计中,还可以在引物的5’端接上3nt-50nt的人工序列、锚定序列或其他设计序列,按锚定引物多重扩增检测或基因芯片检测原理进行其他的扩增检测反应。所述试剂盒还可包括核酸探针。所述核酸探针的序列选自如seqidno:4-seqidno:26所示的的正向和反向互补序列,或者2条之间的seqidno:1的特异性序列或其反向互补的特异性序列;或如seqidno:27-seqidno:34所示序列,或者2条之间的seqidno:2的特异性序列或其反向互补的特异性序列;或如seqidno:35-seqidno:40所示的正向和反向互补序列,或者2条之间的seqidno:3的特异性序列或其反向互补的特异性序列。核酸探针可以用地高辛(dig)、荧光素或者放射性同位素等进行所有核苷酸或特定核苷酸,如dig-dutp,的掺入法标记;也可以用dig、荧光素、报告基团、荧光淬灭基团或放射性同位素等进行末端修饰标记,例如探针的5’端标记fam、hex、vic等,3’端标记淬灭基团tamra等;进行直接对对虾东方病毒或其他变异的同源病毒的基因组核酸进行独立的原位杂交或斑点杂交检测,也可与实时定量pcr技术联合,进行taqman探针或beacon探针的荧光定量pcr检测。所述试剂盒的检测方法包括但不限于常规聚合酶链式反应(pcr)、恒温对流pcr、套氏pcr、实时荧光pcr、恒温对流实时荧光pcr、数字pcr、斑点杂交、原位杂交、环介导等温扩增(lamp)、滚环扩增技术(rca)、单引物等温扩增、依赖解旋酶的等温扩增技术(hda)、交叉引物扩增技术、核酸快递等温检测放大技术;也可包括但不限于同时对1株或1种以上对虾东方病毒与其同源种或变异种的多重pcr、多重实时荧光pcr、基因芯片、芯片检测;还可以包括但不限于同时对包含对虾东方病毒及其同源种或变异种的多重pcr、多重实时荧光pcr、基因芯片、芯片检测。上述检测方法,按照其方法的需要,可以对相应的方法的操作和实验室管理进行能力认证,开展对包括对虾东方病毒及其同源种或变异种的商业化检测服务,或对商业化种苗培育、进出境检疫、产地检疫等开展对包括对虾东方病毒及其同源种或变异种的检测;也可以将相应的试剂和工具进行组装,装配成试剂盒的形式进行对包括对虾东方病毒及其同源种或变异种检测的试剂盒的商业化销售或应用服务;也可以在上述检测方法的基础上研发配套设备,或将已有的配套设备的检测范围扩大覆盖对虾东方病毒及其同源种或变异种,进行对包括对虾东方病毒及其同源种或变异种在内的检测设备的商业化销售或应用服务。所述试剂盒是采用免疫反应进行检测的试剂盒。进一步的,上述试剂盒以(a)如seqidno:1,seqidno:2或seqidno:3所示的核酸序列的互补序列,或其≥70%同源的核酸序列的互补序列翻译的多肽或蛋白质的全部或部分为抗原;或(b)(a)的抗体为检测物。所述抗原可以是病毒株整体、裂解成分、衣壳、多肽、基因工程蛋白或多肽。所述抗体可以为多克隆抗体、杂交瘤细胞、单克隆抗体或单链抗体。所述抗原或抗体可以按照现有技术的方法进行制备。所述试剂盒,可采用竞争性、间接或夹心型免疫反应也可采用固体支持物或免疫沉淀法。所述对虾东方病毒的抗体或抗原性片段可采用现有技术进行标记,如荧光标记、化学发光标记、放射性标记或酶标记。扩增探针信号可以采用生物素和亲和素的方法,酶标记和介导的免疫试验,如elisa检验。如,一种检测对虾东方病毒抗原的试剂盒,试剂盒中含有:固相抗对虾东方病毒抗体包被的孔板、封闭液、小鼠二抗、二次抗体酶结合物、tmb显色液、显色终止液、组织裂解液、浓缩洗液、样品稀释液、阳性参考品和阴性参考品。其中,抗对虾东方病毒抗体包被的孔板预先采用抗布尼亚病毒抗体进行包被;封闭液为人血清白蛋白;样品稀释液为pbs;显色终止液为浓硫酸;浓缩洗液为吐温-20;二次抗体酶结合物为辣根过氧化物酶-羊抗鼠igm酶结合物。本发明的保护范围也包上述对虾东方病毒的抗血清、多克隆抗体、杂交瘤细胞、单克隆抗体或单链抗体。所述抗血清、多克隆抗体、杂交瘤细胞和单克隆抗体以对虾东方病毒的病毒株,病毒株的裂解成份、病毒株的基因工程蛋白或多肽为免疫原按照现有技术的方法进行制备。如,所述抗对虾东方病毒的抗体为多克隆抗体,可以将灭活的对虾东方病毒株或其特定抗原片段,如:l、gn、gc、n的抗原蛋白与适用的佐剂,如:弗氏完全佐剂混合后施予动物,如,鼠、兔、禽(蛋)、猪、山羊、牛、水产动物以进行初次免疫,经适当时间间隔后,视需要以灭活后的病毒液及适当的佐剂施予二次免疫、甚至三次免疫或更多次免疫,以提高抗体效价。经适当时间间隔后,采集免疫动物的血清,即抗血清,进一步分离后获得对虾东方病毒的多克隆抗体。又如,所述抗对虾东方病毒的抗体为单克隆抗体,可将浓缩的对虾东方病毒株或其特定抗原片段,如,l、gn、gc、n的抗原蛋白施予动物,如,小鼠,视需要可添加适当的佐剂,如:弗氏完全佐剂以进行初次免疫。经适当时间间隔后,视需要以灭活的病毒(或抗原蛋白)及适当的佐剂施与二次免疫。经适当时间间隔后,采集免疫动物的血清,用以评估适合用以采集脾脏细胞的小鼠。从所述适用的小鼠采集脾脏细胞与骨髓瘤细胞,如:fo细胞株、ns细胞株以peg进行细胞融合。从融合细胞中筛选出具分泌能力的杂交瘤细胞株后,得到融合细胞系,该融合细胞系可分泌抗对虾东方病毒的单克隆抗体。本发明具有以下优点:本发明的对虾东方病毒是一种新的布尼亚病毒,可感染中国对虾;提供的基于该布尼亚病毒新种的试剂盒可对其进行检测。为甲壳类水产布尼亚病毒的检测与防控提供新的可能。生物保藏信息对虾东方病毒(orientaviruspenaei),保藏编号为cctccno.v201864,保藏单位:中国典型培养物保藏中心(cctcc),保藏地址:中国武汉武汉大学保藏中心,保藏日期:2018年12月06日。附图说明图1为对虾东方病毒的电镜图片;图2为对虾东方病毒的系统发育进化树;图3为中国对虾淋巴器官病理分析电镜图片,淋巴器官中可见病毒包涵体;图4为中国对虾鳃丝病理分析电镜图片,鳃丝中可见病毒包涵体;图5为检测对虾东方病毒的1%琼脂糖凝胶电泳图。具体实施方式下面结合实施例和附图对本发明做进一步说明,但本发明不受下述实施例的限制。实施例1对虾东方病毒的分离、纯化和鉴定1.1分离和纯化(1)取2-3g对虾头胸甲加入到离心管中,预冷的tnep缓冲液和200μlpmsf异丙醇溶液,冰浴下用匀浆器10,000rpm匀浆10s;(2)匀浆液于高速离心机4℃下10,000g离心30min,保留上清;(3)依次使用0.45μm和0.22μm孔径的滤器过滤掉组织碎片、细菌和其他杂质;将上清于超速离心机4℃下100,000g离心180min,用tn缓冲液浸泡并缓慢摇动,使沉淀悬浮,吸取10μl用于电镜观察;(4)根据tem扫描结果,于超速离心管中自上而下铺设蔗糖浓度(w/v)为20%、31.5%、43%、54.5%、66%浓度梯度,待4℃过夜放置后,加入上清进行4℃130,000g离心170min,收集甘油和54.5%蔗糖密度的样品,脱糖后,获得病毒,暂时命名为对虾东方病毒(orientaviruspenaei);(5)病毒序列的全基因组扩增:在通过高通量测序获得部分新型布尼亚病毒序列的基础上,在此基础上设计引物对全基因组进行长片段的扩增和验证,然后采用smarterracecdnaamplificationkit扩增s片段的3’末端和5’末端的以获得全基因组序列,如seqidno:1-seqidno:3所示。表1对虾东方病毒基因组测序引物引物名称序列5'3'l-1fccgggtgtgttctaatgaatctacl-1rcatggatgaaggaaatgggtgl-2fatattgggacgccccctcl-2rcgactctgatggagactacctgttl-3fctgtttcctccggggtatctcl-3rcgatcagtattgtagcagtgccttl-4fcctagtgtgttattcacacaagcattl-4rgttcaagctgcctgaggaacatl-5fctatgaaaccatgcatgtaaccagl-5rtttgaggtacatgaaatacgtgtgttl-6facataagtaccctacatagtttaggttcacl-6rtcacagagtggacatagccttagaal-7fgacattcagcttattactatcgaggal-7rcttatgactatgagctgagggagagl-8ftcatcctcctcatggaaagatactcl-8rggtgttgtagtgttgcattggaal-9fagatgcccttgttaccccgal-9ragccatgcttcagtcaattcagtm-1fgtaatcaatgatcgtttagttggtcttm-1ratctttacagtgggattggaggttm-2ftccaagtccttaacatagaagcagtcm-2rtatggattaggggcaacattgacm-3fccaggagaaacagtgtggattgm-3rgcttgctgccaacaatgctm-4fttggggtgatggaataggttgm-4rcgggagctacaagtctgccats-1facagtggtaagaaggcagacaacs-1rgagagcccagggtgataaacas-2fgacacacagatacacgcacacatacs-2ratgccatgggccttggatas-3ftgaaggaagtggcagcagagts-3rtaacaatcagaatgcttatggtttg1.2种属鉴定对虾东方病毒电镜图片如图1所示,该病毒呈球形,直径80-120nm。根据布尼亚病毒目各个科(属)病毒rna依赖的rna聚合酶(rna-dependentrnapolymerase,rdrp)的氨基酸序列,利用muscle软件进行比对,利用mega软件构建最大相似度进化树(图2),统发育进化树显示该病毒与布尼亚病毒目白纤病毒科(phenuiviridae)的亲缘关系最近。选择白纤病毒科中已知毒株的rdrp序列再次构建最大相似度进化树,显示该病毒属于布尼亚病毒目白纤病毒科(phenuiviridae)中分类地位介于帕西病毒属和白蛉病毒属的一个新的种。对其进行了保藏,保藏编号为cctccno.v201864。实施例2对虾东方病毒对中国对虾的侵染取发病中国对虾样品制备匀浆液,经无菌过滤和超速离心后,通过反向灌肠或肌肉注射对其进行人工感染实验,感染对虾10天内的死亡率可达100%,主要症状表现为活力降低,肝胰腺变浅。感染对虾东方病毒的中国对虾的淋巴器官电镜图片如图4所示,鳃丝病理电镜图片如图5所示。由图片可知,中国对虾的淋巴器官和鳃丝中均可见对虾东方病毒的病毒粒子。实施例3检测对虾东方病毒的pcr试剂盒检测对虾东方病毒的pcr试剂盒包括以下成分:成分浓度extaqbuffer(无mgcl2)10×mgcl225mmdntp每种2.5mml-2613f10μml-3509r10μml-2688f10μml-3030r10μmextaq酶5u/μl无菌双蒸水-表2检测对虾东方病毒的引物序列及扩增片段:参照以下方法检测:将中国对虾的疑似感染组织匀浆提取后,提取rna,调节rna浓度至500ng/μl,再反转录为cdna,然后利用如表2所示的引物进行套式pcr扩增。具体的反应体系与条件如下:配制除taqdna聚合酶以外的大体积预混物,分装保存于-20℃。检测前,吸取无酶预混液,按比例加入相应体积taqdna聚合酶,混匀后分装24μl/管。第一轮扩增程序:94℃变性2min;94℃20s、53℃30s、72℃60s,30个循环;72℃延伸7min。第二轮扩增程序为:94℃变性2min;94℃20s、55℃30s、72℃30s,30个循环;72℃延伸7min。表3第一轮pcr扩增体系成分体积(μl)10×extaqbuffer(无mgcl2)2.5mgcl2(25mm)2dntp(2.5mmeach)2l-2613f(10μm)1l-3509r(10μm)1extaq酶(5u/μl)0.1无菌双蒸水15.4模板:cdna1表4第二轮pcr扩增体系成分体积(μl)10×extaqbuffer(无mgcl2)2.5mgcl2(25mm)2dntp(2.5mmeach)2l-2688f(10μm)1l-3030r(10μm)1extaq酶(5u/μl)0.1无菌双蒸水15.4模板:第一轮pcr产物1将pcr产物以1%琼脂糖凝胶进行电泳,结果如图5所示。图5上图中泳道3和4可以观察到897nt的初次扩增条带。图5下图中可以观察到343nt的初次、二次扩增条带。图5下图中,泳道3-10为感染该病毒的病虾的扩增条带。序列表<110>中国水产科学研究院黄海水产研究所<120>基于对虾东方病毒基因组序列的检测方法及其应用<130>20181115<160>40<170>patentinversion3.5<210>1<211>6317<212>rna<213>orientaviruspenaei<400>1acacaaagacggguguuguaguguugcauuggaaaaaucaagguuuuaaucucucaauua60uggaagaguggcuaaguguguuggagucuaacaacacaggugcuuauguacacaacccuu120cugucaucuuugagcccacagacagcccugagcuauugacuuacgaaaucaagaacccag180acuccauuggccuugugacucaaguggagauugaguuugaugauaggccagaugagggca240cggggaguacauuagccacucaaguccucucagugcagaggguuaggacauuucuucaug300auuucaccuauucucacauaucccacaguacagaugugcgcuuggacacaguuuucacuc360caauggggaauagaggugaucaucucaccccugacguuauucugagagaugguaacaaga420uucuuguuguugaguuugccacaacacgagguggggacgcagcacuagaaagguccuuca480ggacuaaaacagccgcuuaugacuaugagcugagggagagggcugagaaaaggcauucga540aucacuucgacucucagguguucuauggcaucauugugacaaaugagcugaaggugugca600guaaccugccucugacagaagggcaggugaaugagcuggucuacagauauaggauggcaa660uagacauucagacugaguugucuggagcauggggaguaucuuuccaugaggaggaugaug720aaaugucaaagacaacugcagaagucaagaccauucuuaagaacauucccuuaagcuucu780cauuugaugguaaguacauaaacaaggaaguauaugacaacucauucggagcgccugaca840cagauuaccugcagaaggugauuggacgacugaugacagaggguagagcugaagccauga900agagcggggcagagaugaucuucugugaugagagcaaaacaugggguggugagacaguga960gccguaauuccucugagugcuuugaggcaaucaagcaauaugaaggagagcugaugaaag1020gggauaggcagcacagcaguacaaaggcaauuguccagcugccagcuuggacugcaauaa1080gagaagaguucccgggcuccacaucuguccuaaguuuugacauuggggagcacgcgucag1140caacacaugcuguguggaagucugccauguccgcuguggaggcuugggggggguucaaag1200acgaugacgagucagagauaaugaugacugaagaagaagaucaggaucaucacaagccgc1260acagguccaaguaucacagaguggacauagccuuagaagaccacguaagggccagcuuag1320cugugaauggugucuucggcaaggaauucagacggaaugaguuuguggcagcucggagaa1380gugaaaagcggaagccuuucucucuaacuguugacugcucuuguguggaccuuuuccucg1440auaguaauaagcugaaugucgagcuagacacuccgauucaugguaacuuaguucaauuga1500uuuccucaucuacugagaugcauucccaagaugagcaucagaggaaguguaucagcaagu1560ucaugaagucucaguuggggguguauacaucaaucaugacugauguggcgacagagcugu1620gugucagccugaagcagcacgugaagagaaaccaaaugauaauuaagaagcuuagauacu1680auccuguuuacaugcuaauuuacccuacugacucauccaagcacauauucgucucacucc1740uggcugaaagagggaaagugucagugugggacacgacaggguguuucaaaaccagcaugu1800cgaaugacaaggucguguucacugacuuuguuucuuuuaacaccucgaaauuggcuaacc1860ugugcaagcuagagucagucauguauaauacaucaauguucugggcagaggagaacggug1920aggcaagcuucauuggggaugaaaucaugccaugguauggcaagaccucuagaacucagg1980aggaggucuggaagaugaucucucucuguuugcucguggccaugucugacaaaaggcaug2040uugaagagcagcuauccuuguacagguacaugaacauggaggccaugacgucuuucccaa2100gggugccgaagccauacaagcugcugaagaaaaugaccuugguuccgagauccaagcuug2160agcucuggauaaucaagaaacauuugagguacaugaaauacguguguuccggccaucaug2220ccauuaagaugaagggagacacaggagacaccacuuggcauaaccuuaucaaucccuaca2280cguuguuuccacucaccuccaucuccucaaacgugaaccuaaacuauguaggguacuuau2340guaauaaggaggagucagcugagggaaacacagcggccaacaucuaugagaagauucuag2400cucuggaggaugaacuaccagaugucaacacguuccucggccuggaagacccugaugauc2460cuggguaccaugaauucucuccaagucuucuaaaggcagcaguugacaauauaaaggaca2520gauuuagaaggaccuuuggaagcucaugggaggccaacauagacaauaagauucugagag2580cucuuggcagcauaacuuuagaggaaauggccacgcugaaagcuuccucgaacuucucuc2640cugaauacuaugcuuugaagccaggagaugcguauaaaagaaaaaaagccauccaaggag2700ugagagacuugauggauggggagucuacaaugauguaugagguccugaguaagugccucu2760caaaaguggagcaagauggcugcaugcacauagauauuuucaagaagccucaacacggag2820gagauagggagauuuauguuauggacaugucaucaaggguuguccagcuaggguuagaga2880ccauugcuagaaccuacuguucuuucauagacucagaggcaaugacucacccgaagagca2940aguucaagcugccugaggaacaugaaaggaaugcagagaaacgcuuggguacucauguga3000cguucugucagagcgcagaugcuaggaaguggagucaagggcaccauguugcuaaguuca3060ugcagaugcuaugcaggcuaacuccuaguuacauacaugguuucauagugagagcgugug3120cgcucuggacgaaaaggcgaauaaugaucucaccagcucugauaaagguguuuugcgaaa3180gcacuagcuuuacuucaggcaacgauaucgugcagaggauguaugauggguauuuaggua3240aaggaacagagaaguggauaacccagggcauguccuacauacagguuggcaguggcauga3300ugcaaggcauccuucacuacacuucgucccugcugcacucaaucuuucaagaguuucuaa3360ggagcuugcugagaucucauuauagggcacgaagccugcugauaggggaacccaugauca3420cugugggccagagcucugaugacuccuaucucaugguuacuuuuagaacgagggauucug3480aaacaugggcugaggcagcacuugagucaucaaugguccaccaccucaaguccacauuga3540guagauuccuggggaucuaugacucugagaagacugcuagcauguugaugucugucaugg3600aguuccuuagcgaguacauggaaggcgccagccuacauagggcaacugugaagucagugu3660uugcuugcuuaucagugagugagcaugagagccugucuucucgccaggaagaaaugagca3720cccuccugacuaaggugguugaggauggaggaagcauuaaucuggccucucauugucaau3780ugggccaggcucucuuacacuauaggcuauuggggucaucaguguccccugcuuucgauc3840aguauuguagcagugccuucacauccagagacccugccuuagguuacuuccugauggaca3900acccauacgcugcugggcugcuugguuuuaaauacaaccuguggaaugcuugugugaaua3960acacacuagggaagaaguauaagcagauguuaaucaacccccugagacauaggccucugg4020agucaacaacaucaggaacuuuacuucacagucaucauauugcuugggggaaucggaaaa4080aaugggaaaggcugguuagagcuuuggaguuacccgagaauuggagggaucugauagagg4140aggauccaguaauccuauauagaaaggcccaaacagagagagacuuggagcuuagguugg4200ccgagaaaaugcacucaccuggaguuugcucaucucugaguaagggaaaugcugucacua4260gaauaauagcuggcucagucuacauacugaccaggaaugucuuaaccaucguuggaggga4320gaacaaaggugaguuugcucaaagcuauacucgaagagagggacgggguugacccgcucu4380ccugggaggaagagucccuucuguuuccgagugcacuagaauaccacucaguugcugcag4440cccugagaagacuggacucgagagcagggguguucaagaaacauaaggaacgaaggagga4500gguccaacauacagauaaccgacucugauggagacuaccuguuucaaccagaugucauuu4560uagccuggagaugguuuaacauugacaggaugcacgcugcuguucgggugaaggagagga4620uguaucagaaauuaaggcaggcgcugccgugggugagagauaccccggaggaaacagugg4680cgaacagcccguuugagaaucaaauccagcugcacacguucuugacuaagacaaccauga4740agaggagggaggugcaucugcucggaacagaagucacucaaagaggcggaaucucaagcc4800uaaugacugccauuuaccagaaccauuucccuggguuccacuugcaguucuugaaggaug4860aggaggcugcaucacaaucuucugacuacucaaaacuaucucacuucauucacaugacuc4920accuguccccauacucugaugacuacaagcagcaguugauucucaauagacucgccucua4980gcccccagauugaguucaaggagaaguuugggaggaguaggaggaacugccuagccauca5040uccagaagucguugucauugaaugucguagaucuguuagaccacauaaccuucaguuaua5100agaugggaguugucggggguuuuguuaagagacagacaagcacuguuguuaaugggaaag5160ugaaguacacuggcgugggaacguggguuggauccauggacggugucaacaugcgaauug5220ucaucaaugggacaguggaauccaauagagugacuuccauagucgugucaaacccucgag5280caauguacacgcagaccuuugggaggcuacugcagacauggaugaaggaaauggguguua5340cugaaggggacccacacguuaacccaaaugugauagcuuacuacagaaaugggcacauau5400aucagggcggcaggggaggaggagggggcgucccaauauaucacucggggcuaggccuug5460agggauuugauccugccuuggucaaauugguggauauccaggugaggaacaacacgguga5520gacuuguugcagacuauggccugaagcaccucagcaagguaacuauacugagcuacacca5580ccagagacaaggaugcgucauugauggugacuaggucugauuucucagagauaaagccag5640cuaugcaguacugguucucuaacacaucauggccccacgagguagcacagggagugguga5700ccagggcuucuaagggggaacucaugccaaauguugaucacagggaguugcaauuuugga5760ugcaagcacuauuuaagaaugcagcagagaggaaagggcuccuugaggagacaauggagc5820ucccaauggggucuaugacuucgacagugguuccugauguguaugaugauuucugggauc5880uggucaccuuugaugagccacaagagcuagaauguuuuggagaggcugugccuucguggg5940aggacuucgacuuugagguggcuuuuggaggggccaacgucaccaacuauguuccuagug6000uccuccgcacucacccacuucuagaucaaaugcucagcaggcugaucagggacuucuccu6060ucggagccaugagcaaguugguuagggagaagguuggaaccccagccaugcuucagucaa6120uucaguuguuguccuacuugcugggaguggauucaaaugagauagccaucccagacuggg6180auuuagcgucaaguggcgcagagcagugggaccuuuaagauguagcauguugguucauuu6240gauagguuaauucaaugguugguggguacuaaaaaaauaguuauguagauucauuagaac6300acacccggucuuugugu6317<210>2<211>2978<212>rna<213>orientaviruspenaei<400>2acacaaagacggcgcaucaagugagaauucaguagugaaguucauuugguuuagucuuug60cgauauguuuguuuuuuugaucacugcgggcuugcugccaacaaugcugguuggcaacca120acuaucucccgggaguaugcucacagcuggggugggcgaaugugcccagaguccaccgag180auucccauuugcagacccuucuaucaagggugcaaagaagauacaguggaucaaccuauu240ccaucaccccaaacccugcuauuaugagguuacuuccagccaaagauguuuugaugagcu300cggcggaacuggagcaagguguaacaacagcagaggaacugugaugguugaugacauaaa360cugccaaguggaggcugaaguugggucacuuagugacucaaaggacaucugugcagugaa420uggacauauucugcacacuugugcugagaccaggacuggacuugagugggugaggugggc480uuucauguaccgugaaggaaggaagguccgauuccuagacacguugcacaagagagugaa540gcacaauguagcaggcucagacaguuacacuugcacaggaaaucagacagaggggugcaa600uggagacuuagccuacugcucgaacaaugacugugaaguugccucaaccugcuucugcac660aaccaacaugagagaggugaccucccugauugucgauggacaagagauagugccaacaug720cuggggagaaucucuggugaggaucaaucgagagaucauaggcaauguaggggcauucca780gccaugcacagacugcaaguuccagugcaccgaugagggugucaagguuuccacauucau840gcaggaagcugugccuggaaccauaugcaagaaccccuauugcguccacaucaugguuca900guaugaagcuucaauaguccugccccuggacaugagagucucaacugaagagaugacuau960cacccuauggauuaggggcaacauugacccguucgucaacaaaaugacuugcguguauga1020gcaauccugcgcucucauuuccugccaucuuugccuggagagagcucucaauccacacug1080uuucuccugggcccacuggcuguuagucaugguuuuaauccuggucagugcagcugccau1140ccgcaggaucaucaugcuccugcucugccucaagugggcggugauuacaugcuggcacug1200ucugaaguggauccaccaaagggcuucuuccuugaggaggggcaaggcuaugcaggacga1260ugaccgggccagauuaacucaaggacgggcaggagcugcuaacagguucagggccuaccu1320ggccauggcccugcugucccucggucacugcugcuccacaucagucaccucuauagcuga1380cauccaggacuguucgcaagguacagacggaguugagacuugugccuuugaccacggagu1440ggaucugcaugugaguccaauuggccaggagaguuguuacuuccucaaggaccccaaagg1500ggagcacauggaucaccuucguauuaagaccgugagcguucagcugggcugugaaaagca1560gucccuguacuucgucccuagggcaguugguaagugcgccaguaucagucacuguucaac1620uguugauggcugcucaucaaaagcgugccucgaguucggcaucaaugacacaaaggcuga1680cuggacaguagaggcagagcacuacgguuggucuagaugccguggaaucccaggcugcgc1740ugcaaauggguguuucuacugugaugaugggugccucugguggagagaguacuucaccaa1800ccccaagauggagguguucgagguggucaggugcccgaccuggaucuuuacagugggauu1860ggagguuguaguguccaacguuacuacgcccauaacccuaucuccaggagcuacaaaagc1920uguuggcucagugaggcugagcuuggaggcucucagcgugccaccugagccauuauuggg1980agacugcuucuauguuaaggacuuggagacaaagauugggccuugcaacgagagaggguc2040gcugagcccuggaagaguuggggagcugcaaugcccaucuaaggagucagcaagaagagu2100ugacaaaacuuguuuugcgaaugacgcuauugucagguccacugugagcucggcaggggu2160gucaugucacuuuuccuuaguggacccagagaagguuggaaccgaccucccguacaaagg2220caauggcuucacaauccugucagggaaagaaggaauaguggcucauaguaccuccaccgc2280uuugaugucccucaauauacagcuggggcgcuugcugauccaaaaaagagcugccaggua2340caaaugucaugcucaguuugucaaacugaccggguguuacucuugcauggcuggugccac2400ccugacuuuauuagugggcuccaccucugaugcuucagaggcugugcucaacugcccuga2460cgccaacuacaccacaauuguuguugcagacaagcuggagaagagcgugaccagcacaau2520gcaccugacugaaucacagauagacaugaagugcgagauuguguguccgaguagcaggac2580auuugucgaagucuccgguaaucucuuguacaucccggccucggacccugaaacuaggac2640caugucgguggacguucacaacaauggagcaucauucacauggaacccccucgggagcua2700caagucugccaucuuguauggggcaauugcuauagcugcacucauccuccuuucuauucu2760ccuuccucuccuacagaccucugugaaaucuaagaccaacuaaacgaucauugauuacga2820cgggccgaaaggcucgcauccgcuaacuaacuguuuggcagagcuauccggcucaacaag2880ugaggccacugacauguugaguuucaacguguggaagauucuugcuuugcaaucuuaaca2940cacuucacuauauuugucauaugcgccggucuuugugu2978<210>3<211>1164<212>rna<213>orientaviruspenaei<400>3acacaaagacgagggaucuauagucaauaacagaguauucuuaccuuguucaacugacaa60cuuuucaacauggccucuagugcugagcuuagugaggcugcgaaguucauucagaacauc120gccgucggugaguuuacgaucuucguagccgacauggaugcguucaucgcagacauccag180uuccagggcuuugacccuaaggucauuguugcagccuugaugaagaaggagagcgaugca240aacacgcucaaggaggacaucugcaagaugguggccauucugugugagaggggaaccaag300cucaacaagaucauggguaggucuucuccggagggaguuucucggaucagggcccucaag360gauaaauauggccuuguggacagugguaagaaggcagacaacaucacccuugcucggguu420gccuucugccuuccccaguucacaugcucguacauggcggucugucugaaccccgcgguu480ccauggaccagccuagacgaggggaccuaugccuauccaaggcccauggcaugcagugcc540uucgcaaaccugcuagaggcgaccgauacggagaugaucaacugucaccuguacugggca600guccauuuuaacaagcugaucaacccaucagccaacaaguccaugaaggaaguggcagca660gagugccuuaaguuugccaugauaggggcaaacucgacccacaucccagccgcuaagaag720aagucaaucagggaagcccugguccuaucugcgccaacaaucaaguucuacucugauaag780uucuugagcuugaccgccuagaaaggcucucacauuaguggggccagacggugacacaca840aacacacuaaauagacaagcacgacaugagaacaacacagacgagacacaaacaacucac900aagacacacagauacacgcacacauacacacacacacacacaauccgagguccugugugu960gacuuugaaaugauggagcugcacacucacucgcucuccuccugcuugugcucaugcuga1020ccccccaauguguggcacauucaagcauucugauugcuuguuccucggucuuuuauagac1080ucaaggagcucauggucuguucgucaagaauauuaugaggcaaaccauaagcauucugau1140uguuauaucccucggucuuugugu1164<210>4<211>23<212>dna<213>artificialsequence<220><223>l-12f<400>4ggtgttgtagtgttgcattggaa23<210>5<211>25<212>dna<213>artificialsequence<220><223>l-718r<400>5tcatcctcctcatggaaagatactc25<210>6<211>25<212>dna<213>artificialsequence<220><223>l-496f<400>6cttatgactatgagctgagggagag25<210>7<211>26<212>dna<213>artificialsequence<220><223>l-1460r<400>7gacattcagcttattactatcgagga26<210>8<211>25<212>dna<213>artificialsequence<220><223>l-1274f<400>8tcacagagtggacatagccttagaa25<210>9<211>30<212>dna<213>artificialsequence<220><223>l-2342r<400>9acataagtaccctacatagtttaggttcac30<210>10<211>20<212>dna<213>artificialsequence<220><223>l-2613f<400>10acgctgaaagcttcctcgaa20<210>11<211>20<212>dna<213>artificialsequence<220><223>l-3509r<400>11tgactcaagtgctgcctcag20<210>12<211>21<212>dna<213>artificialsequence<220><223>l-2688f<400>12gccatccaaggagtgagagac21<210>13<211>20<212>dna<213>artificialsequence<220><223>l-3011r<400>13acttcctagcatctgcgctc20<210>14<211>20<212>dna<213>artificialsequence<220><223>l-3030r<400>14acttcctagcatctgcgctc20<210>15<211>26<212>dna<213>artificialsequence<220><223>l-3085f<400>15tttgaggtacatgaaatacgtgtgtt26<210>16<211>24<212>dna<213>artificialsequence<220><223>l-3108r<400>16ctatgaaaccatgtatgtaactag24<210>17<211>22<212>dna<213>artificialsequence<220><223>l-2942f<400>17gttcaagctgcctgaggaacat22<210>18<211>26<212>dna<213>artificialsequence<220><223>l-3970r<400>18cctagtgtgttattcacacaagcatt26<210>19<211>24<212>dna<213>artificialsequence<220><223>l-3836f<400>19cgatcagtattgtagcagtgcctt24<210>20<211>21<212>dna<213>artificialsequence<220><223>l-4677r<400>20ctgtttcctccggggtatctc21<210>21<211>24<212>dna<213>artificialsequence<220><223>l-4520f<400>21cgactctgatggagactacctgtt24<210>22<211>18<212>dna<213>artificialsequence<220><223>l-5439r<400>22atattgggacgccccctc18<210>23<211>21<212>dna<213>artificialsequence<220><223>l-5317f<400>23catggatgaaggaaatgggtg21<210>24<211>24<212>dna<213>artificialsequence<220><223>l-6308r<400>24ccgggtgtgttctaatgaatctac24<210>25<211>23<212>dna<213>artificialsequence<220><223>l-6104f<400>25agccatgcttcagtcaattcagt23<210>26<211>20<212>dna<213>artificialsequence<220><223>l-9r<400>26agatgcccttgttaccccga20<210>27<211>19<212>dna<213>artificialsequence<220><223>m-90f<400>27gcttgctgccaacaatgct19<210>28<211>22<212>dna<213>artificialsequence<220><223>m-1090r<400>28ccaggagaaacagtgtggattg22<210>29<211>23<212>dna<213>artificialsequence<220><223>m-966f<400>29tatggattaggggcaacattgac23<210>30<211>26<212>dna<213>artificialsequence<220><223>m-2007r<400>30tccaagtccttaacatagaagcagtc26<210>31<211>24<212>dna<213>artificialsequence<220><223>m-1844f<400>31atctttacagtgggattggaggtt24<210>32<211>27<212>dna<213>artificialsequence<220><223>m-2818r<400>32gtaatcaatgatcgtttagttggtctt27<210>33<211>21<212>dna<213>artificialsequence<220><223>m-2692f<400>33cgggagctacaagtctgccat21<210>34<211>21<212>dna<213>artificialsequence<220><223>m-252r<400>34ttggggtgatggaataggttg21<210>35<211>23<212>dna<213>artificialsequence<220><223>s-380f<400>35acagtggtaagaaggcagacaac23<210>36<211>21<212>dna<213>artificialsequence<220><223>s-1120r<400>36cctcataatattcttgacgaa21<210>37<211>21<212>dna<213>artificialsequence<220><223>s-644f<400>37tgaaggaagtggcagcagagt21<210>38<211>25<212>dna<213>artificialsequence<220><223>s-1145r<400>38taacaatcagaatgcttatggtttg25<210>39<211>25<212>dna<213>artificialsequence<220><223>s-903f<400>39gacacacagatacacgcacacatac25<210>40<211>19<212>dna<213>artificialsequence<220><223>s-532r<400>40atgccatgggccttggata19当前第1页12
相关技术
网友询问留言
已有0条留言
- 还没有人留言评论。精彩留言会获得点赞!
1