胶质母细胞瘤危险分层模型及其在临床预后评估的应用的制作方法
本发明属于生物技术领域,具体涉及一种胶质母细胞瘤患者预后风险分层模型和临床预后评估。
背景技术:
胶质母细胞瘤(glioblastomas,gbm)是神经胶质瘤中危害最严重的亚类(whoiv),占所有脑胶质瘤发病人数的54%。gbm的1年生存率仅为1/3,其5年死亡率达到95%,在全身肿瘤死亡率排名中位列第三。近年研究发现1p/19q缺失、idh1和tert突变、h3f3a突变和mgmt启动子甲基化等基因分子事件对神经胶质瘤分型诊断和药物治疗具有参考指导价值,但目前这些生物标记物仅对低级别的神经胶质瘤(whoii、iii)的分型诊断具有指导价值,对危险级别最高的胶质母细胞瘤,临床评估还没有可行的风险分类生物标记物。
近年随着高通量技术应用的普及,发现全基因组范围内的联合生物标记物成为可能,具有很高的临床转化研究潜力。目前生物分子和病理特征可以区分神经胶质瘤的基本亚型,可评估低级别的神经胶质瘤亚型的临床预后。但针对母细胞瘤患者的临床预后,临床医生急需找到一个敏感性高、通用性好和特异性强的预后标志物,来帮助实现病人健康管理预警。基于全基因组甲基化芯片数据,研究发现两个预后模型(3和6-cpg),但是由于数据平台异质性、样本数量限制、snp及模型不包含非cpg探针等因素,使得该模型仅适用于较小的特征人群,并不能广泛适用于母细胞瘤患者。
现有技术cn106324248a公开了aqp1一种全新的人神经胶质瘤预后指标,所述指标包括:对特定细胞模型及临床样品的aqp1表达量进行测定以确定其与细胞增殖及侵袭力、β-catenin表达、肿瘤病理分级及组织学分级、总生存期(os)及无进展生存期(pfs)等因素之间的关系,同时将aqp1表达量与患者预后效果相比较,以确定其作为患者辅助治疗中潜在预后标记物和药物靶标的重要性。
现有技术cn102408478a公开了一种人少突神经胶质瘤标志物map2蛋白及其用途,该map2蛋白可用作人少突神经胶质瘤的蛋白标志物,尤其能用作区分临床上具有化疗敏感的,1p等位基因杂合性缺失的人少突神经胶质瘤的蛋白标志物。该map2蛋白可用于准确预测具有90%的1p等位基因杂合性缺失,可用于少突神经胶质瘤1p缺失样品化疗敏感的机制研究,以及通过决策树模型等方式预测1p等位基因杂合性缺失的病人的化疗敏感性,为诊断和预后以及治疗提供靶点。
神经母细胞瘤的发病机理仍不明确,因此目前急需新的临床预后标志物来实现胶质母细胞瘤风险分类,以实现胶质母细胞瘤的精准治疗。
技术实现要素:
针对于现有技术的不足,本发明提供一种建立胶质母细胞瘤患者预后分层模型的方法。
同时,本发明还提供一种胶质母细胞瘤患者预后风险评估系统。
本发明还提供一种探针组,所述的探针组能够用于检测患者基因甲基化位点甲基化率,以预测胶质母细胞瘤患者预后风险,为病人预后及健康管理提供指导依据。
一种用于提供信息以建立胶质母细胞瘤预后风险分层模型的方法,所述的方法包括如下步骤:
(1)、在tcga数据库中选择胶质母细胞瘤的患者基因的甲基化位点的信息,利用单因素cox模型获得探针风险系数和相应p值,筛选出胶质母细胞瘤显著相关的甲基化探针组a,检测胶质母细胞瘤患者的基因甲基化位点的甲基化率,并且获得患者的生存时间信息;
(2)、从癌症相关体细胞突变位点数据库获得肿瘤相关基因和从omim获得胶质母细胞瘤相关基因,通过对比获得交集基因,获得上述交集基因对应的探针组b;
(3)、取探针组a与b的交集,去除交集中探针风险系数的异常值,获得胶质母细胞瘤相关的甲基化探针组c,利用探针组c中的各探针的由单因素cox模型得到的风险系数构建加权方程,形成风险分层模型;
(4)、将步骤(1)中tcga数据库中选择胶质母细胞瘤的患者样本的甲基化率带入风险模型给出模型值,排序后,取中位数即判断阈值,小于中位数的样本归为预后高风险组,大于等于中位数的样本归为预后低风险组。
所述的步骤(1)中的基因甲基化位点的甲基化率是通过现有技术中常用的检测方法进行检测得到,例如,利用高通量测序法检测甲基化位点的甲基化率。
优选的,所述的步骤(3)中利用boxplot方法去除交集中探针风险系数的异常值。
所述的探针组a、探针组b、探针组c的序列信息通过测序芯片的注释信息获得。
在本发明的一个具体实施方式中,在建立胶质母细胞瘤预后风险分层模型的方法中,所述的步骤(1)中,在tcga数据库中选择所有胶质母细胞瘤患者产生于illuminahumanmethylation450array平台的甲基化位点的信息。
在本发明的一个具体实施方式中,在建立胶质母细胞瘤预后风险分层模型的方法中,所述的步骤(3)中,所获得与胶质母细胞瘤相关的甲基化探针组c为111个探针,所述的111个探针的序列为seqidno:1-143,甲基化探针上的甲基化位点为:(cg07464524,cg09912841,cg11426662,cg25560327,cg24550026,cg09692771,cg13681847,cg16483490,cg20623601,cg24046888,cg25124739,cg02970696,cg05726118,cg07556134,cg12508343,cg18011163,cg24105729,cg00202441,cg01483139,cg02647878,cg02968741,cg08660876,cg09760963,cg18016365,cg07862423,cg13593479,cg17621438,cg04488476,cg09013068,cg12798992,cg00376553,cg01900030,cg02204205,cg02316066,cg05231706,cg05898452,cg06586813,cg08300570,cg11139102,cg11274371,cg15774495,cg18071865,cg21517389,cg21808635,cg22281380,cg23104539,cg25819027,cg27006764,cg01981354,cg24820936,cg24828864,cg17028652,cg12041266,cg16036142,cg21558509,cg12836863,cg26740195,cg04352704,cg00645339,cg04025675,cg05896902,cg09093388,cg10088041,cg19114050,cg24328539,cg25908973,cg26796135,cg00291054,cg01769813,cg06330323,cg06788514,cg06846976,cg07549381,cg07644807,cg08779982,cg08936056,cg26819590,cg06270615,cg06788790,cg07780782,cg09967973,cg10792831,cg17833746,cg18711081,cg19636308,cg25330422,cg26111030,cg05724997,cg14623989,cg14889643,cg17602451,cg21793948,cg23707289,cg26277730,cg06346838,cg08165462,cg12400781,cg13368085,cg13422347,cg14559388,cg24156854,cg00876694,cg01118078,cg02872693,cg16626405,cg16810054,cg17204562,cg21859623,cg26204638,cg19810954,cg26237628),表1所示为所述的甲基化位点信息,与所述的各位点相对应的风险系数为(0.014927198,0.009302326,0.006020697,0.009157401,0.008749802,0.012970721,0.014429021,0.006045153,0.009023346,0.009275152,0.009737098,0.008170105,0.001622246,0.009220806,0.010443604,0.004156609,0.012499717,0.007488961,0.006822309,0.002885804,0.008027898,0.013948959,0.014111999,0.006864881,0.01252689,0.011177283,0.016294921,0.008895632,0.004353162,0.012590295,0.017726048,0.012372908,0.005900229,0.007700913,0.008767012,0.012798623,0.005853128,0.004294287,0.006162904,0.002009918,0.006125767,0.00682865,0.015017776,0.004951881,0.011720749,0.013794977,0.01066099,0.004364032,0.01508118,0.016285863,0.016756867,0.001413917,0.004810579,0.010117525,0.007691855,0.010026947,0.014483368,0.009266095,0.004050633,0.001293449,0.001814271,0.003696474,0.00912117,0.01406671,0.003540681,0.006263445,0.002057924,0.017852857,0.001876769,0.014084826,0.005609474,0.0030262,0.012472544,0.004660221,0.009329499,0.003301556,0.003058808,0.009565001,0.016910849,0.007900183,0.014573945,0.004452798,0.012019655,0.00621091,0.006219062,0.001239102,0.00920269,0.017427142,0.009637463,0.003555173,0.003643939,0.014972487,0.015380087,0.01463735,0.010651933,0.010760626,0.017870972,0.006777926,0.004264396,0.013939902,0.01175698,0.016077534,0.01293449,0.005075972,0.007195489,0.002834175,0.003175653,0.003180182,0.011711691,0.011032359,0.017309391),所述的胶质母细胞瘤预后的风险分层模型为:
y=coe*x
coe=(0.014927198,0.009302326,...,0.017309391),x=(v1,v2,...,v111)t。
表1甲基化位点信息
所述的甲基化位点是利用illuminahumanmethylation450array甲基化芯片获得的,所述的cg07464524,cg09912841……cg19810954,cg26237628为甲基化位点在illuminahumanmethylation450array甲基化芯片中的编号,所述的甲基化位点的信息可从illuminahumanmethylation450array注释信息中获取。
在本发明的一个具体实施方式中,在建立胶质母细胞瘤预后风险分层模型的方法中,所述的步骤(4)中,所述的阈值为0.4385954。
本申请的发明人创造性的提出一种建立癌症预后预测模型的方法,解决了现有技术中由于数据平台异质性、样本数量限制、snp及模型不包含非cpg探针等因素造成的模型适用范围小的不足的问题。
本发明提供一种胶质母细胞瘤患者预后风险评估方法,所述的评估方法包括如下步骤:
(1)获取待检测患者dna的111个甲基化位点的甲基化率,所述的111个甲基化位点为(cg07464524,cg09912841,cg11426662,cg25560327,cg24550026,cg09692771,cg13681847,cg16483490,cg20623601,cg24046888,cg25124739,cg02970696,cg05726118,cg07556134,cg12508343,cg18011163,cg24105729,cg00202441,cg01483139,cg02647878,cg02968741,cg08660876,cg09760963,cg18016365,cg07862423,cg13593479,cg17621438,cg04488476,cg09013068,cg12798992,cg00376553,cg01900030,cg02204205,cg02316066,cg05231706,cg05898452,cg06586813,cg08300570,cg11139102,cg11274371,cg15774495,cg18071865,cg21517389,cg21808635,cg22281380,cg23104539,cg25819027,cg27006764,cg01981354,cg24820936,cg24828864,cg17028652,cg12041266,cg16036142,cg21558509,cg12836863,cg26740195,cg04352704,cg00645339,cg04025675,cg05896902,cg09093388,cg10088041,cg19114050,cg24328539,cg25908973,cg26796135,cg00291054,cg01769813,cg06330323,cg06788514,cg06846976,cg07549381,cg07644807,cg08779982,cg08936056,cg26819590,cg06270615,cg06788790,cg07780782,cg09967973,cg10792831,cg17833746,cg18711081,cg19636308,cg25330422,cg26111030,cg05724997,cg14623989,cg14889643,cg17602451,cg21793948,cg23707289,cg26277730,cg06346838,cg08165462,cg12400781,cg13368085,cg13422347,cg14559388,cg24156854,cg00876694,cg01118078,cg02872693,cg16626405,cg16810054,cg17204562,cg21859623,cg26204638,cg19810954,cg26237628),所述的甲基化位点信息如表1所述;
(2)将步骤(1)中获取的甲基化率带入风险模型中,计算模型值,所述的风险模型为:
y=coe*x
coe=(0.014927198,0.009302326,...,0.017309391),x=(v1,v2,...,v111)t。
将得到的模型值与阈值0.4385954相比较,模型值小于阈值的患者为胶质母细胞瘤高风险组,模型值大于等于阈值的患者为胶质母细胞瘤低风险组,所述的coe=(0.014927198,0.009302326,0.006020697,0.009157401,0.008749802,0.012970721,0.014429021,0.006045153,0.009023346,0.009275152,0.009737098,0.008170105,0.001622246,0.009220806,0.010443604,0.004156609,0.012499717,0.007488961,0.006822309,0.002885804,0.008027898,0.013948959,0.014111999,0.006864881,0.01252689,0.011177283,0.016294921,0.008895632,0.004353162,0.012590295,0.017726048,0.012372908,0.005900229,0.007700913,0.008767012,0.012798623,0.005853128,0.004294287,0.006162904,0.002009918,0.006125767,0.00682865,0.015017776,0.004951881,0.011720749,0.013794977,0.01066099,0.004364032,0.01508118,0.016285863,0.016756867,0.001413917,0.004810579,0.010117525,0.007691855,0.010026947,0.014483368,0.009266095,0.004050633,0.001293449,0.001814271,0.003696474,0.00912117,0.01406671,0.003540681,0.006263445,0.002057924,0.017852857,0.001876769,0.014084826,0.005609474,0.0030262,0.012472544,0.004660221,0.009329499,0.003301556,0.003058808,0.009565001,0.016910849,0.007900183,0.014573945,0.004452798,0.012019655,0.00621091,0.006219062,0.001239102,0.00920269,0.017427142,0.009637463,0.003555173,0.003643939,0.014972487,0.015380087,0.01463735,0.010651933,0.010760626,0.017870972,0.006777926,0.004264396,0.013939902,0.01175698,0.016077534,0.01293449,0.005075972,0.007195489,0.002834175,0.003175653,0.003180182,0.011711691,0.011032359,0.017309391),所述的x为与所述的风险系数相对应的甲基化位点的甲基化率,所述的步骤(1)中获得111个甲基化位点的甲基化率分别带入v1,v1,…….v111中。
优选的,所述的胶质母细胞瘤患者预后风险评估方法的步骤(1)中,包括取待检测患者的组织样本,提取dna,检测步骤(1)中获得的dna的111个甲基化位点的甲基化率。
优选的,所述的胶质母细胞瘤患者预后风险评估方法的步骤(1)中,取待检测患者的血液样本。
所述的胶质母细胞瘤患者预后风险评估方法的步骤(1)中,提取待检测患者的组织样本中的dna是通过现有技术中常用dna提取方法进行,例如,利用商品化的dna提取试剂盒提取待检测患者的组织样本中的dna。
所述的胶质母细胞瘤患者预后风险评估方法的步骤(2)中,甲基化率是通过现有技术中常用的检测方法进行检测得到,例如,利用高通量测序法检测甲基化位点的甲基化率。
一种用于胶质母细胞瘤患者临床预后风险评估系统,所述的预后风险评估系统包括数据输入模块、模型计算模块和结果输出模块,所述的模型计算模块通过风险分层模型计算获得胶质母细胞瘤患者的模型值,所述的风险分层模型为:
y=coe*x
coe=(0.014927198,0.009302326,...,0.017309391),x=(v1,v2,...,v111)t,
所述的coe=(0.014927198,0.009302326,0.006020697,0.009157401,0.008749802,0.012970721,0.014429021,0.006045153,0.009023346,0.009275152,0.009737098,0.008170105,0.001622246,0.009220806,0.010443604,0.004156609,0.012499717,0.007488961,0.006822309,0.002885804,0.008027898,0.013948959,0.014111999,0.006864881,0.01252689,0.011177283,0.016294921,0.008895632,0.004353162,0.012590295,0.017726048,0.012372908,0.005900229,0.007700913,0.008767012,0.012798623,0.005853128,0.004294287,0.006162904,0.002009918,0.006125767,0.00682865,0.015017776,0.004951881,0.011720749,0.013794977,0.01066099,0.004364032,0.01508118,0.016285863,0.016756867,0.001413917,0.004810579,0.010117525,0.007691855,0.010026947,0.014483368,0.009266095,0.004050633,0.001293449,0.001814271,0.003696474,0.00912117,0.01406671,0.003540681,0.006263445,0.002057924,0.017852857,0.001876769,0.014084826,0.005609474,0.0030262,0.012472544,0.004660221,0.009329499,0.003301556,0.003058808,0.009565001,0.016910849,0.007900183,0.014573945,0.004452798,0.012019655,0.00621091,0.006219062,0.001239102,0.00920269,0.017427142,0.009637463,0.003555173,0.003643939,0.014972487,0.015380087,0.01463735,0.010651933,0.010760626,0.017870972,0.006777926,0.004264396,0.013939902,0.01175698,0.016077534,0.01293449,0.005075972,0.007195489,0.002834175,0.003175653,0.003180182,0.011711691,0.011032359,0.017309391);所述的x中v1,v1,…….v111为与所述的风险系数相对应的甲基化位点(cg07464524,cg09912841,cg11426662,cg25560327,cg24550026,cg09692771,cg13681847,cg16483490,cg20623601,cg24046888,cg25124739,cg02970696,cg05726118,cg07556134,cg12508343,cg18011163,cg24105729,cg00202441,cg01483139,cg02647878,cg02968741,cg08660876,cg09760963,cg18016365,cg07862423,cg13593479,cg17621438,cg04488476,cg09013068,cg12798992,cg00376553,cg01900030,cg02204205,cg02316066,cg05231706,cg05898452,cg06586813,cg08300570,cg11139102,cg11274371,cg15774495,cg18071865,cg21517389,cg21808635,cg22281380,cg23104539,cg25819027,cg27006764,cg01981354,cg24820936,cg24828864,cg17028652,cg12041266,cg16036142,cg21558509,cg12836863,cg26740195,cg04352704,cg00645339,cg04025675,cg05896902,cg09093388,cg10088041,cg19114050,cg24328539,cg25908973,cg26796135,cg00291054,cg01769813,cg06330323,cg06788514,cg06846976,cg07549381,cg07644807,cg08779982,cg08936056,cg26819590,cg06270615,cg06788790,cg07780782,cg09967973,cg10792831,cg17833746,cg18711081,cg19636308,cg25330422,cg26111030,cg05724997,cg14623989,cg14889643,cg17602451,cg21793948,cg23707289,cg26277730,cg06346838,cg08165462,cg12400781,cg13368085,cg13422347,cg14559388,cg24156854,cg00876694,cg01118078,cg02872693,cg16626405,cg16810054,cg17204562,cg21859623,cg26204638,cg19810954,cg26237628)的甲基化率,所述的甲基化位点信息如表1所述,所述的阈值为0.4385954。
所述的风险分层模型是加权方程,coe为风险系数,x为待检测患者基因的111个甲基化位点的甲基化率,所述的风险模型是以基因甲基化位点的风险系数乘以该位点的甲基化率加权计算,得模型值。
所述的数据输入模块,用于将患者的基因甲基化位点的甲基化率值输入模型计算模块。
所述的模型计算模块还包括将所述的模型计算模块中获得的模型值与阈值相比,模型值小于阈值为高风险组,模型值大于阈值为低风险组。
所述的结果输出模块用于输出判断结果。
在本发明的一个具体实施方式中,将测得的患者的基因甲基化位点的甲基化率利用数据输入模块输入到模型计算模块,模型计算模块中的风险分层模型以患者的基因甲基化位点的甲基化率为变量进行加权计算,得到模型值,模型值与阈值相比后,得到患者预后风险,模型值小于阈值为高风险组,模型值大于阈值为低风险组,然后通过结果输出模块输出判断结果。
一种用于检测胶质母细胞瘤患者预后风险的探针组,所述的探针组为seqidno:1-143中的两种或两种以上的组合。
一种用于检测胶质母细胞瘤患者预后风险的探针组,所述的探针组为seqidno:1-143的组合。
序列为seqidno:1-143的探针在建立胶质母细胞瘤危险分层模型或者在制备胶质母细胞瘤患者临床预后风险评估产品中的应用。
优选的,所述的胶质母细胞瘤患者临床预后风险评估产品包括试剂盒、芯片及装置。
本发明所述的tcga数据库是指癌症和肿瘤基因组图谱数据库,是由美国国家癌症研究所及国家人类基因组研究所联合建立,其获取网址为https://cancergenome.nih.gov/。
本发明所述的癌症相关体细胞突变位点数据库是指cosmic数据库,其获取网址为https://cancer.sanger.ac.uk/cosmic/。
本发明所述的omim是指人类基因和遗传紊乱的数据库,其获取网址为http://omim.org/。
附图说明
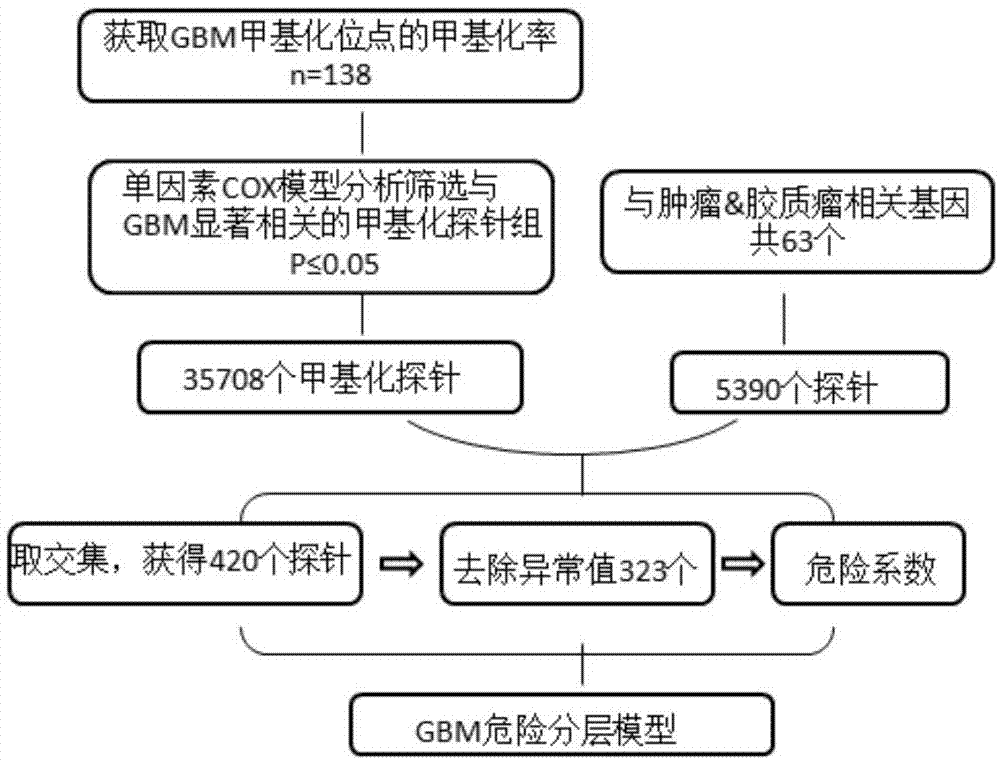
图1为本发明实施例中危险分层模型构建过程示意图;
图2为利用生存曲线法对建立模型的138名患者的数据验证本发明提供的系统中的风险分层模型结果图;
图3为利用生存曲线法对geo数据库中62名患者的数据验证本发明提供的系统中的风险分层模型结果图。
具体实施方式
下面结合附图和实施例对本发明做进一步详细说明。所述是对本发明的解释,而不是限定。
实施例1一种用于预测胶质母细胞瘤患者预后风险的系统
一种用于预测胶质母细胞瘤患者预后风险的系统,该系统中包括数据输入模块、模型计算模块和结果输出模块。
数据输入模块用于将测得的患者的基因甲基化位点的甲基化率值输入模型计算模块。
模型计算模块中包括判断胶质母细胞瘤患者预后的风险分层模型,风险分层模型为:
y=coe*x
coe=(0.014927198,0.009302326,...,0.017309391),x=(v1,v2,...,v111)t,
coe=(0.014927198,0.009302326,0.006020697,0.009157401,0.008749802,0.012970721,0.014429021,0.006045153,0.009023346,0.009275152,0.009737098,0.008170105,0.001622246,0.009220806,0.010443604,0.004156609,0.012499717,0.007488961,0.006822309,0.002885804,0.008027898,0.013948959,0.014111999,0.006864881,0.01252689,0.011177283,0.016294921,0.008895632,0.004353162,0.012590295,0.017726048,0.012372908,0.005900229,0.007700913,0.008767012,0.012798623,0.005853128,0.004294287,0.006162904,0.002009918,0.006125767,0.00682865,0.015017776,0.004951881,0.011720749,0.013794977,0.01066099,0.004364032,0.01508118,0.016285863,0.016756867,0.001413917,0.004810579,0.010117525,0.007691855,0.010026947,0.014483368,0.009266095,0.004050633,0.001293449,0.001814271,0.003696474,0.00912117,0.01406671,0.003540681,0.006263445,0.002057924,0.017852857,0.001876769,0.014084826,0.005609474,0.0030262,0.012472544,0.004660221,0.009329499,0.003301556,0.003058808,0.009565001,0.016910849,0.007900183,0.014573945,0.004452798,0.012019655,0.00621091,0.006219062,0.001239102,0.00920269,0.017427142,0.009637463,0.003555173,0.003643939,0.014972487,0.015380087,0.01463735,0.010651933,0.010760626,0.017870972,0.006777926,0.004264396,0.013939902,0.01175698,0.016077534,0.01293449,0.005075972,0.007195489,0.002834175,0.003175653,0.003180182,0.011711691,0.011032359,0.017309391);x中v1,v1,…….v111为与风险系数相对应的甲基化位点(cg07464524,cg09912841,cg11426662,cg25560327,cg24550026,cg09692771,cg13681847,cg16483490,cg20623601,cg24046888,cg25124739,cg02970696,cg05726118,cg07556134,cg12508343,cg18011163,cg24105729,cg00202441,cg01483139,cg02647878,cg02968741,cg08660876,cg09760963,cg18016365,cg07862423,cg13593479,cg17621438,cg04488476,cg09013068,cg12798992,cg00376553,cg01900030,cg02204205,cg02316066,cg05231706,cg05898452,cg06586813,cg08300570,cg11139102,cg11274371,cg15774495,cg18071865,cg21517389,cg21808635,cg22281380,cg23104539,cg25819027,cg27006764,cg01981354,cg24820936,cg24828864,cg17028652,cg12041266,cg16036142,cg21558509,cg12836863,cg26740195,cg04352704,cg00645339,cg04025675,cg05896902,cg09093388,cg10088041,cg19114050,cg24328539,cg25908973,cg26796135,cg00291054,cg01769813,cg06330323,cg06788514,cg06846976,cg07549381,cg07644807,cg08779982,cg08936056,cg26819590,cg06270615,cg06788790,cg07780782,cg09967973,cg10792831,cg17833746,cg18711081,cg19636308,cg25330422,cg26111030,cg05724997,cg14623989,cg14889643,cg17602451,cg21793948,cg23707289,cg26277730,cg06346838,cg08165462,cg12400781,cg13368085,cg13422347,cg14559388,cg24156854,cg00876694,cg01118078,cg02872693,cg16626405,cg16810054,cg17204562,cg21859623,cg26204638,cg19810954,cg26237628)的甲基化率,甲基化位点信息如表1所述,通过数据输入模块输入的基因甲基化位点的甲基化率值与风险系数进行加权计算,得模型值,模型值与阈值相比,小于阈值的样本归为预后高风险组,大于等于阈值的样本归为预后低风险组,阈值为0.4385954。
结果输出模块用于将上述判断结果输出。
上述风险分层模型是用如下方法得到的:
(1)检测胶质母细胞瘤患者的基因甲基化位点的甲基化率,并且跟踪患者的生存时间信息,共得到138例样本信息;
(2)利用单因素cox模型分析筛选与胶质母细胞瘤显著相关的甲基化探针组,获得各探针风险系数和相应p值,筛选出胶质母细胞瘤显著相关的35708个甲基化探针。从cosmic数据库获得肿瘤相关基因和从omim获得胶质瘤相关基因,通过对比获得63个交集基因,即同时与肿瘤和胶质瘤相关的基因集。对照illuminahumanmethylation450array注释信息,获得对应的探针集,总计5390个探针;
(3)通过将以上获得的两个探针集取交集,获得420个探针。进一步对探针风险系数采用boxplot方法去除异常值,获得最终111个甲基化探针组,该甲基化探针组即seqidno:1-143的组合,该甲基化探针上有甲基化位点即上述111个甲基化位点。利用各探针风险系数构建加权方程,形成风险模型y=coe*x;
(4)分别将检测得到的138个样本中每个样本的111个甲基化位点的甲基化率带入风险模型给出模型值,排序后,取中位数即判断阈值,中位数为0.4385954。
图1为危险分层模型构建过程示意图。
在预测患者的胶质母细胞瘤预后风险过程中,检测患者上述111个甲基化位点的甲基化率,然后带入上述风险模型进行加权计算,得到模型值,模型值小于中位数的样本归为预后高风险组,模型值大于等于中位数的样本归为预后低风险组。
利用生存曲线法对138名患者进行验证,验证结果如图2所示。图2所示的结果表明,生存曲线法验证显示,利用本系统中的风险分层模型对患者分组效果显著。
实施例2
对实施例1提供的系统中的风险模型进行验证。取geo数据库中gse60274数据集,该数据集是胶质母细胞瘤患者于illuminahumanmethylation450array平台产生的甲基化芯片信息,该数据有患者完整生存信息,该数据集中共有62名患者的数据,具体数据见(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse60274)。应用风险分层模型,将gse60274数据集中62名患者每个患者的上述111甲基化位点的甲基化率代入实施例1中的风险分层模型进行加权计算,得到的模型值与阈值相比,将患者按高低风险分组,结果如附图3所示。
图3的结果显示,将geo数据库中gse60274数据集中62名患者每个患者的上述111甲基化位点的甲基化率带入风险分层模型进行加权计算后,得到的模型值与阈值相比后区分高风险组和低风险组,通过生存曲线法验证显示分组效果显著。
序列表
<110>中国人民解放军总医院
<120>胶质母细胞瘤危险分层模型及其在临床预后评估的应用
<130>20181018
<160>143
<170>patentinversion3.5
<210>1
<211>50
<212>dna
<213>human
<400>1
aactaaaaacaaaacaaaaactacaaaacaaacccaaaaccaaacaaaca50
<210>2
<211>50
<212>dna
<213>human
<400>2
aactaaaaacgaaacgaaaactacaaaacgaacccaaaaccaaacaaacg50
<210>3
<211>50
<212>dna
<213>human
<400>3
aaaaacaaacactcctaaaacttcaaacaccaattttcttacracaaaac50
<210>4
<211>50
<212>dna
<213>human
<400>4
aacaaaatctacaactacaaaaaataacaacacacacaaacatactcaca50
<210>5
<211>50
<212>dna
<213>human
<400>5
aacaaaatctacgactacaaaaaataacgacacgcgcaaacgtactcacg50
<210>6
<211>50
<212>dna
<213>human
<400>6
catttcccaacaaacaccacaaacaaaatctacaactacaaaaaataaca50
<210>7
<211>50
<212>dna
<213>human
<400>7
catttcccgacgaacaccgcaaacaaaatctacgactacaaaaaataacg50
<210>8
<211>50
<212>dna
<213>human
<400>8
taatttaccaatatttttaacttcctactaaactaaaaactacttatttc50
<210>9
<211>50
<212>dna
<213>human
<400>9
tacattctttctactctaccatctaaaatcacctaaactattactaaacc50
<210>10
<211>50
<212>dna
<213>human
<400>10
aaaaaaaaatcaatctactaatcaactattaaaatttttactaaacaccc50
<210>11
<211>50
<212>dna
<213>human
<400>11
ctaactataacaactattctcctccaacaacttcctctctctcctaatac50
<210>12
<211>50
<212>dna
<213>human
<400>12
aaaacctaacaataatcttaacaaatatataaaatatcaactcctcctac50
<210>13
<211>50
<212>dna
<213>human
<400>13
aaaaacttatatattttacaactacratacctatcraaaatataacaatc50
<210>14
<211>50
<212>dna
<213>human
<400>14
cactataaaaactacctacraaacrccttaaaaaccrctatataaactac50
<210>15
<211>50
<212>dna
<213>human
<400>15
acaaaataaaactacataataacaaataaactaccacccacctaaatcca50
<210>16
<211>50
<212>dna
<213>human
<400>16
acgaaataaaactacgtaataacgaataaactaccgcccgcctaaatccg50
<210>17
<211>50
<212>dna
<213>human
<400>17
attacacccacaaaaaaaacccatatcctcttttaccaaattaataaaac50
<210>18
<211>50
<212>dna
<213>human
<400>18
ctaaccctacacccaacaaaaaaactaaaaacaaaaactaaatacctcca50
<210>19
<211>50
<212>dna
<213>human
<400>19
ctaaccctacgcccgacgaaaaaactaaaaacgaaaactaaatacctccg50
<210>20
<211>50
<212>dna
<213>human
<400>20
acacaaaaaaaaatcataaaaaatactaatccctacatacaaacacactc50
<210>21
<211>50
<212>dna
<213>human
<400>21
atatccaccaaacaacaaaaaaaccaccaaaaactaataataaacaccca50
<210>22
<211>50
<212>dna
<213>human
<400>22
atatccgccaaacaacgaaaaaaccgccgaaaactaataataaacacccg50
<210>23
<211>50
<212>dna
<213>human
<400>23
atttataaatacctaataccacacaaaaatcaaactctcaaaattcttac50
<210>24
<211>50
<212>dna
<213>human
<400>24
aaattctcaaatacrtcctaaatttcctctatacctttaataactctccc50
<210>25
<211>50
<212>dna
<213>human
<400>25
aaaatatacctataaaacrcaactttacaaaaattataatcaatatcaac50
<210>26
<211>50
<212>dna
<213>human
<400>26
caaaaccactactaaaaaaaacactaaccaacatacaaaaaacaaaaaca50
<210>27
<211>50
<212>dna
<213>human
<400>27
cgaaaccgctactaaaaaaaacgctaaccaacgtacaaaaaacgaaaacg50
<210>28
<211>50
<212>dna
<213>human
<400>28
taaaatactccatatcctccraataatcaaaaacaaactactatttccac50
<210>29
<211>50
<212>dna
<213>human
<400>29
cttcaaaacctcaaacctcccattcaaccaaccattatcrccrataaaac50
<210>30
<211>50
<212>dna
<213>human
<400>30
aaaaaactaacaccccacacaataacatcttccaaaaaatcatacatcca50
<210>31
<211>50
<212>dna
<213>human
<400>31
aaaaaactaacaccccgcacgataacatcttccaaaaaatcatacgtccg50
<210>32
<211>50
<212>dna
<213>human
<400>32
ttcraaaattactacccaacaaccractaaattacattccttaaatcttc50
<210>33
<211>50
<212>dna
<213>human
<400>33
tatactccrctttccctcattacaatcaaaaaacattttctacaaaaaac50
<210>34
<211>50
<212>dna
<213>human
<400>34
ctaaccccaaactatcractctaaaaatactaaaactaaaacctaccttc50
<210>35
<211>50
<212>dna
<213>human
<400>35
actaacaacrctacctaaatttaattttcccttcrtatatcactatcttc50
<210>36
<211>50
<212>dna
<213>human
<400>36
aaacraaataaaactcataaaatactttaaatctcctcctcctcctcttc50
<210>37
<211>50
<212>dna
<213>human
<400>37
ataaataaaaacacccatcacttatcatattaatttccaacaatcctttc50
<210>38
<211>50
<212>dna
<213>human
<400>38
atctacccatactaacttccaaaaacctctatatattcctatatataaac50
<210>39
<211>50
<212>dna
<213>human
<400>39
tcacttcccctctcaaacttctcaaactcaaataaaattctactcctaca50
<210>40
<211>50
<212>dna
<213>human
<400>40
tcgcttcccctctcaaacttctcgaactcaaataaaattctactcctacg50
<210>41
<211>50
<212>dna
<213>human
<400>41
attctctaacttaactaaaaatctattcaccttaaataactcctccattc50
<210>42
<211>50
<212>dna
<213>human
<400>42
tcrcttataaattatctttcraaacaaatccaatttatcctttcactaac50
<210>43
<211>50
<212>dna
<213>human
<400>43
acaccaaaaccttataaaaccacaaacaaacctcaaatctataactacca50
<210>44
<211>50
<212>dna
<213>human
<400>44
acgccaaaaccttataaaaccacgaacaaacctcgaatctataactaccg50
<210>45
<211>50
<212>dna
<213>human
<400>45
acctaatatactcttaaaaaaccaactaacaaaaccctataaatcttaac50
<210>46
<211>50
<212>dna
<213>human
<400>46
aacaaaaataaataacaaacaacctaaacctcaaacttttaaaactaaac50
<210>47
<211>50
<212>dna
<213>human
<400>47
aaaaatcrtcaattttaaacrctttctcctaaaaatccacatattcctac50
<210>48
<211>50
<212>dna
<213>human
<400>48
aaatcacaaaacctattcaaacttctccccaaaaccccaaaacaacatac50
<210>49
<211>50
<212>dna
<213>human
<400>49
ccacaatataaaaataacctaaaactaaatatctaaatcctatcaaaacc50
<210>50
<211>50
<212>dna
<213>human
<400>50
ccaaacacttttcactttcaacataaactacatttattttattttccttc50
<210>51
<211>50
<212>dna
<213>human
<400>51
ctactacrcccaaaataaaaaatctaaactacrcaattaatactttatac50
<210>52
<211>50
<212>dna
<213>human
<400>52
aaacttttaaaactaaacrccaaaaccttataaaaccacraacaaacctc50
<210>53
<211>50
<212>dna
<213>human
<400>53
caaaatattaactacrcaatccaaatcctttatcctaaacrcaacaaaac50
<210>54
<211>50
<212>dna
<213>human
<400>54
cacaacaaaaatctcaaaaccacaaccaaaaaaacaacaaaaaacataca50
<210>55
<211>50
<212>dna
<213>human
<400>55
cgcaacaaaaatctcaaaaccacaaccaaaaaaacgacgaaaaacatacg50
<210>56
<211>50
<212>dna
<213>human
<400>56
aaacaaatccaatttatcctttcactaacaaaacctcctaaaccacaaca50
<210>57
<211>50
<212>dna
<213>human
<400>57
aaacaaatccaatttatcctttcactaacgaaacctcctaaaccgcgacg50
<210>58
<211>50
<212>dna
<213>human
<400>58
ataaaaaatctaaactacrcaattaatactttatacrtttcaaacaattc50
<210>59
<211>50
<212>dna
<213>human
<400>59
taattcttaaactataaaccttcttaacatcactatcttaccaaattacc50
<210>60
<211>50
<212>dna
<213>human
<400>60
ctttcraaacaaatccaatttatcctttcactaacraaacctcctaaacc50
<210>61
<211>50
<212>dna
<213>human
<400>61
atcaaacaaccctaaacttacataaaatttcaatctcaacttcctcatac50
<210>62
<211>50
<212>dna
<213>human
<400>62
actaaaatttaaacccataaatactacaaaataattaccactatacaacc50
<210>63
<211>50
<212>dna
<213>human
<400>63
tcaaaatctcaccctcttcctctaacaaattttactacctaataattttc50
<210>64
<211>50
<212>dna
<213>human
<400>64
tcccaaactttcctttttatacaacratccrtaacttttctctttacctc50
<210>65
<211>50
<212>dna
<213>human
<400>65
ttaaaaaaaacaactccaaactttaaaactaacaaccaaaaaccatatac50
<210>66
<211>50
<212>dna
<213>human
<400>66
aattataaactatacataccaacaaaactatcaacaaaaatatattaacc50
<210>67
<211>50
<212>dna
<213>human
<400>67
tatcctacaataaacaaacaacaaaaaaaacaaacttaacaaaaaaacca50
<210>68
<211>50
<212>dna
<213>human
<400>68
tatcctacaataaacaaacgacgaaaaaaacaaacttaacgaaaaaaccg50
<210>69
<211>50
<212>dna
<213>human
<400>69
attataaaacccatcttaactcrctcataaaatataaaataccttactcc50
<210>70
<211>50
<212>dna
<213>human
<400>70
ttatcactccracrcatcctctctctactttttaaataaacttttaactc50
<210>71
<211>50
<212>dna
<213>human
<400>71
ataacrtacttaaaacacaaaaaaaacacaacattacrtatactcaaaac50
<210>72
<211>50
<212>dna
<213>human
<400>72
ataataaaaccaacataacaccccaaattaaaaactatcaaaaaacacca50
<210>73
<211>50
<212>dna
<213>human
<400>73
ataataaaaccgacgtaacgccccgaattaaaaactatcgaaaaacgccg50
<210>74
<211>50
<212>dna
<213>human
<400>74
tttaaattctcaaatattcacttaatatatacataaaccattatcacttc50
<210>75
<211>50
<212>dna
<213>human
<400>75
caaatattcacttaatatatacataaaccattatcacttcrtcaaaaatc50
<210>76
<211>50
<212>dna
<213>human
<400>76
taaatctataaccccaaaacttaccaaataaaacacacaccaaacaaaca50
<210>77
<211>50
<212>dna
<213>human
<400>77
taaatctataaccccgaaacttaccgaataaaacgcacgccaaacgaacg50
<210>78
<211>50
<212>dna
<213>human
<400>78
caaataataaaaccaacataacaccccaaattaaaaactatcaaaaaaca50
<210>79
<211>50
<212>dna
<213>human
<400>79
caaataataaaaccgacgtaacgccccgaattaaaaactatcgaaaaacg50
<210>80
<211>50
<212>dna
<213>human
<400>80
tcccatcaaaaactcttctctcaatcctaaaaaacaatataccraaaacc50
<210>81
<211>50
<212>dna
<213>human
<400>81
aatattcacttaatatatacataaaccattatcacttcrtcaaaaatcrc50
<210>82
<211>50
<212>dna
<213>human
<400>82
ccaaaaacaaataataaaaccaacataacaccccaaattaaaaactatca50
<210>83
<211>50
<212>dna
<213>human
<400>83
ccgaaaacaaataataaaaccgacgtaacgccccgaattaaaaactatcg50
<210>84
<211>50
<212>dna
<213>human
<400>84
atacataaaccattatcacttcatcaaaaatcacatcctacccaattaca50
<210>85
<211>50
<212>dna
<213>human
<400>85
atacataaaccattatcacttcgtcaaaaatcgcgtcctacccaattacg50
<210>86
<211>50
<212>dna
<213>human
<400>86
taaaacttcaaaataaaaacaccactttctcactacacttccattttatc50
<210>87
<211>50
<212>dna
<213>human
<400>87
aacctcacctaaccttctccaacatacaaaacracratcttataaatatc50
<210>88
<211>50
<212>dna
<213>human
<400>88
tcctcaaatctacccaaacaacttataactaacactaaaccccatacaca50
<210>89
<211>50
<212>dna
<213>human
<400>89
tcctcaaatctacccaaacaacttataactaacactaaaccccgtacgcg50
<210>90
<211>50
<212>dna
<213>human
<400>90
aactccaaaatcacaaaaaccrcraacacaatcaaaaacatctaaaaacc50
<210>91
<211>50
<212>dna
<213>human
<400>91
cccacracccataaaactcaaaacrtcaaaaacrctaaaactataataac50
<210>92
<211>50
<212>dna
<213>human
<400>92
cractactaactctctaaacaccaaactaacaatcactaaaaactaaacc50
<210>93
<211>50
<212>dna
<213>human
<400>93
atcraactaaattccctcttatatctttcctaaatccttccraaaaaaac50
<210>94
<211>50
<212>dna
<213>human
<400>94
aaaccacattaccatcaccaaaaacaccaaaccaataaaaacaaaaccca50
<210>95
<211>50
<212>dna
<213>human
<400>95
aaaccacattaccgtcaccgaaaacaccaaaccaataaaaacaaaacccg50
<210>96
<211>50
<212>dna
<213>human
<400>96
aaaactcaaacaaaactctatataccacaatcacaatcctttaaacratc50
<210>97
<211>50
<212>dna
<213>human
<400>97
tcaacacaaaccctctatacaaccccaaaaacaataaatcctaatatcca50
<210>98
<211>50
<212>dna
<213>human
<400>98
tcgacacaaaccctctatacaaccccaaaaacgataaatcctaatatccg50
<210>99
<211>50
<212>dna
<213>human
<400>99
ctttaattctacctccttcacttaaaaaacaactctctactaacaaccac50
<210>100
<211>50
<212>dna
<213>human
<400>100
ataaaatttaaatacaaaactccccaaaaacccaaactaaaaataataac50
<210>101
<211>50
<212>dna
<213>human
<400>101
caacttataaaccaccttataaataaataaacaacccacraaattccctc50
<210>102
<211>50
<212>dna
<213>human
<400>102
acaacatcttactcatcttaaaaaaccaaaaatttcttccacacacacca50
<210>103
<211>50
<212>dna
<213>human
<400>103
gcgacgtcttactcatcttaaaaaaccgaaaatttcttccacacacaccg50
<210>104
<211>50
<212>dna
<213>human
<400>104
acatacrtaaacacctcctacaacccactaacraactaaaaaaaattaac50
<210>105
<211>50
<212>dna
<213>human
<400>105
acaaaaaatacaacaactaaaacaaatactcactacrctaaaccaaatac50
<210>106
<211>50
<212>dna
<213>human
<400>106
taaccatattatatcccaaaaaactttttcaacaaacaaaacaactatca50
<210>107
<211>50
<212>dna
<213>human
<400>107
taaccgtattatatcccgaaaaactttttcgacaaacgaaacaactatcg50
<210>108
<211>50
<212>dna
<213>human
<400>108
cratcaaaacaaacraaacaaaaaatcacaacaatcacaacaatcacacc50
<210>109
<211>50
<212>dna
<213>human
<400>109
actataaataacctataacatttacttacaaaaacaaacaaaaaaatacc50
<210>110
<211>50
<212>dna
<213>human
<400>110
aaatactacaaaaaataaaaaccaaacacacaacacaataaaaaacccca50
<210>111
<211>50
<212>dna
<213>human
<400>111
aaatactacgaaaaataaaaaccgaacgcgcgacgcaataaaaaaccccg50
<210>112
<211>50
<212>dna
<213>human
<400>112
atatatatatcrcctaaaccctttctaaccrtatataaaaatatatacac50
<210>113
<211>50
<212>dna
<213>human
<400>113
ctatacaaaccatccacctaaaacaaaactacaaaaaaaacaacatccca50
<210>114
<211>50
<212>dna
<213>human
<400>114
ctatacgaaccatccacctaaaacaaaactacaaaaaaaacaacatcccg50
<210>115
<211>50
<212>dna
<213>human
<400>115
aaaatcatcaccaacctaacaaaaaatcaaataaaccacaaataacacca50
<210>116
<211>50
<212>dna
<213>human
<400>116
aaaatcgtcgccgacctaacgaaaaatcaaataaaccacaaataacaccg50
<210>117
<211>50
<212>dna
<213>human
<400>117
crccracctaacraaaaatcaaataaaccacaaataacaccraactaaac50
<210>118
<211>50
<212>dna
<213>human
<400>118
cactactaaaaacaataaaaaacaacattaacctaaatcttcccccaaca50
<210>119
<211>50
<212>dna
<213>human
<400>119
cactactaaaaacgataaaaaacgacattaacctaaatcttcccccgacg50
<210>120
<211>50
<212>dna
<213>human
<400>120
tcccactactaaaaacaataaaaaacaacattaacctaaatcttccccca50
<210>121
<211>50
<212>dna
<213>human
<400>121
tcccactactaaaaacgataaaaaacgacattaacctaaatcttcccccg50
<210>122
<211>50
<212>dna
<213>human
<400>122
atatataaaaatatatacacacacctacacacacacacattatattacca50
<210>123
<211>50
<212>dna
<213>human
<400>123
gtatataaaaatatatacacgcgcctacacacacacacgttatattaccg50
<210>124
<211>50
<212>dna
<213>human
<400>124
aaatcaaaatcaaaccractaaaccraaactacccacacctaaaatcctc50
<210>125
<211>50
<212>dna
<213>human
<400>125
raaaaccctacccacattctcacrcttaaaaaactattaaaatctacacc50
<210>126
<211>50
<212>dna
<213>human
<400>126
aaaaactttacaaaaattattttcattctcaaccccaactataaaaatac50
<210>127
<211>50
<212>dna
<213>human
<400>127
cattctcaaccccaactataaaaatacraataaaaatataaccraacccc50
<210>128
<211>50
<212>dna
<213>human
<400>128
aaaacccaaaacaaacrcracctcratacttttacaaatcctacaacctc50
<210>129
<211>50
<212>dna
<213>human
<400>129
aatccaaacctactacaaaacctactaaaactaactacaaacccaaaaca50
<210>130
<211>50
<212>dna
<213>human
<400>130
aatccgaacctactacgaaacctactaaaactaactacaaacccgaaacg50
<210>131
<211>50
<212>dna
<213>human
<400>131
aaccatattaaaaacacaccatccctaaaaaaaataactaaaactcaaca50
<210>132
<211>50
<212>dna
<213>human
<400>132
aaccatattaaaaacgcaccatccctaaaaaaaataactaaaactcaacg50
<210>133
<211>50
<212>dna
<213>human
<400>133
tcaattcccaaaaacccttaaaaataaacactaaaaattaaacccaaacc50
<210>134
<211>50
<212>dna
<213>human
<400>134
ctaaaatactcrataaaaaaatcctccaaaaaactactctaaaaacraac50
<210>135
<211>50
<212>dna
<213>human
<400>135
atcaaatcccctaaacttcaaaacacttaaaccttaaataccatccaacc50
<210>136
<211>50
<212>dna
<213>human
<400>136
taaaaaccaaaatccaaactaaaaaactaaaataataaaaccaaaaaccc50
<210>137
<211>50
<212>dna
<213>human
<400>137
aaaacaacttcatctaccaaccataccaacaccaattcaactactaaaca50
<210>138
<211>50
<212>dna
<213>human
<400>138
aaaacaacttcatctaccaaccgtaccaacgccaattcaactactaaacg50
<210>139
<211>50
<212>dna
<213>human
<400>139
tcttccacaaaaaacracaaaaactccaaaaaataaccraaaaccaatac50
<210>140
<211>50
<212>dna
<213>human
<400>140
acracaacatattttaacaaaaattatttactcctaaaaaacccaaaaac50
<210>141
<211>50
<212>dna
<213>human
<400>141
tctaacactataatactaccrataacrtaaacraacatactaaaatactc50
<210>142
<211>50
<212>dna
<213>human
<400>142
taatcratccaactctaaaaaattaataacacatatatctctaatataac50
<210>143
<211>50
<212>dna
<213>human
<400>143
aataaaaaacaaaactcccaatcttctaccctaaacttacctactaaacc50
- 还没有人留言评论。精彩留言会获得点赞!