核酸检测方法与流程
本发明涉及基因工程领域。具体而言,本发明涉及新的基于crispr系统的核酸检测方法。更具体而言,本发明涉及cas12b介导的dna检测方法及相关试剂盒。
背景技术:
快速便携的核酸检测有望在临床诊断和检疫检测中发挥重要作用。crispr核酸酶cas12a和cas13,由于具有ssdna或ssrna反式切割活性已经被开发用于以高灵敏度和特异性快速检测dna和rna。基于crispr-cas13的rna检测平台称为sherlock,基于cas12a的dna检测平台也称作detectr。
开发进一步的具有更高灵敏度和更高特异性的核酸检测平台在本领域具有重要意义。
发明简述
本发明提供一种基于cas12b的核酸检测方法,称为cdetection,能够以比cas12a更高的特异性检测dna,并且具有高达亚渺摩尔(attomolar)级的灵敏度。本发明还提供了增强的cdetection(ecdetection),其能够实现区分仅有单个核苷酸多态性不同的两种靶dna。基于crispr-cas12b的cdetection技术能够提供快速简便的dna检测方法,适用于健康领域和生物技术的各种应用。
附图描述
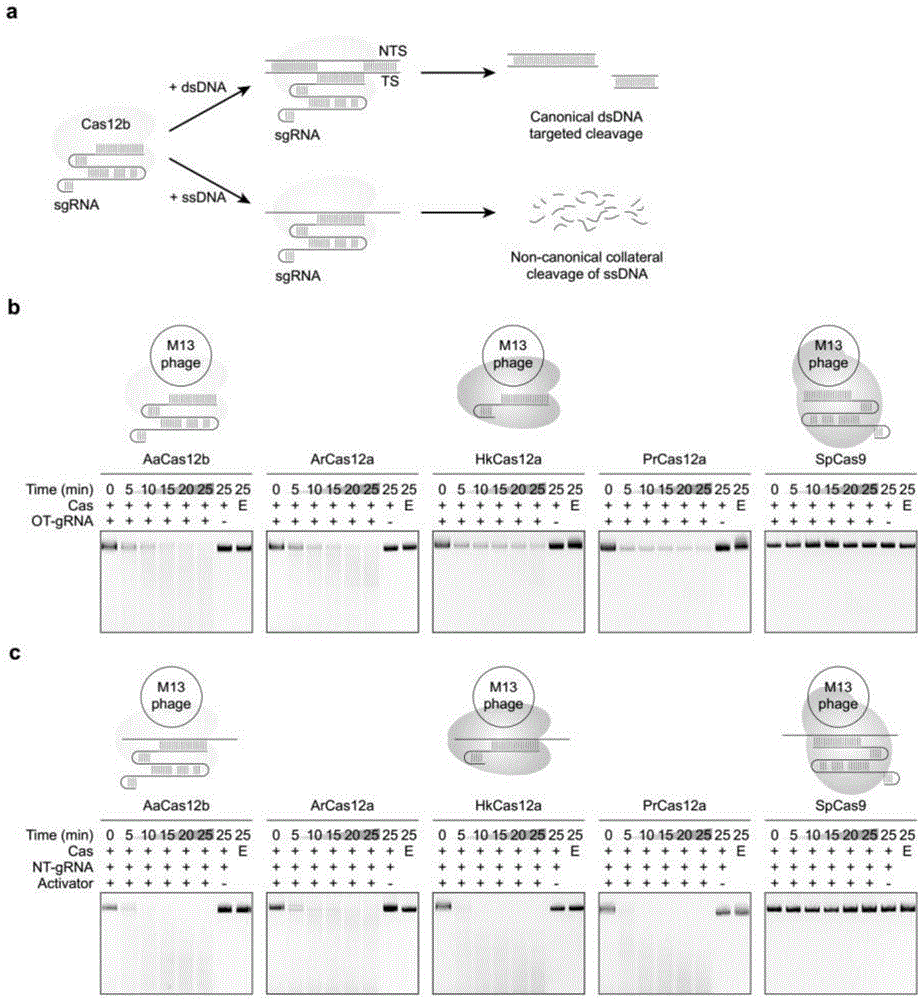
图1.cas12蛋白中保守的靶识别激活非特异性单链dna(ssdna)切割。(a)示意图说明cas12b具有经典的dsdna靶识别和切割,以及非经典的旁路ssdna切割。(b)m13mp18ssdna底物切割时间曲线,其中纯化的aacas12b、arcas12a、hkcas12a、prcas12a和spcas9和与m13噬菌体互补的向导rna(靶grna,ot(ontarget)-grna)结合。(c)m13mp18ssdna底物切割时间过程,其中纯化的aacas12b、arcas12a、hkcas12a、prcas12a和spcas9和与m13噬菌体没有序列同源性的grna(非靶grna,nt(non-target)-grna)及互补ssdna激活物结合。
图2.cas12b的ruvc结构域负责ssdna反式切割。(a-b)m13mp18ssdna底物和puc19dsdna切割时间曲线,其中纯化的wtaacas12b和ruvc催化突变体(r785a或d977a)与靶grna(ot-grna)或非靶grna(nt-grna)结合(a),或与nt-grna及互补ssdna激活物(b)结合,其中非靶grna与m13噬菌体或puc19没有序列同源性。
图3.cas12b介导的反式激活切割对非特异性ssdna的偏好性。(a)aacas12b介导的对含有a、t、g或c核苷酸同聚体的ssdna报告分子的反式切割的核苷酸偏好性。将aacas12b与靶向合成ssdna靶1或靶2的sgrna一起孵育。误差棒表示平均值的标准误差(s.e.m.),n=3。rfu,相对荧光单位。(b)测试多种缓冲液对cas12b介导的反式激活切割的影响。将aacas12b与靶向合成ssdna靶1的sgrna一起孵育。误差棒表示平均值的标准误差(s.e.m.),n=3。(c)使用on-target-ssdna(靶-ssdna或ot-ssdna)、non-target-ssdna(非靶-ssdna或nt-ssdna)、on-target-dsdna(靶-dsdna或ot-dsdna)和non-target-dsdna(非靶-dsdna或nt-dsdna)作为激活物,对aacas12b的反式切割活性分析。误差条表示s.e.m.,n=3。
图4.cas12b介导的dna检测的特异性和灵敏度。(a)使用具有指示的单个错配的ssdna或dsdna激活物分析aacas12b的反式切割活性。误差条表示s.e.m.,n=3。rfu,相对荧光单位。pt,完美配对的靶。mpam,突变的pam。(b)使用具有指示的连续错配的ssdna或dsdna激活物分析aacas12b的反式切割活性。误差棒表示s.e.m.,n=3。(c)比较aacas12b、prcas12a、lbcas12a在区分dsdna方面的特异性,使用合成的hpv16激活物。误差条表示s.e.m.,n=3。(d)使用dsdna激活物比较aac2c1的反式切割活性和预扩增增强的反式切割活性(cdetection)。将aacas12b与靶向靶1合成dsdna的sgrna一起温育。误差条表示s.e.m.,n=3。rpa,重组酶聚合酶扩增。(e)基于aacas12b、prcas12a、lbcas12a的使用rpa预扩增的dna检测所获得的最大荧光信号。将cas12蛋白与靶向合成hpv16dsdna的grna孵育,所述合成hpv16dsdna与背景基因组混合。误差条表示s.e.m.,n=3。(f)基于aacas12b、lbcas12a的使用rpa预扩增的dna检测所获得的荧光时间曲线。将cas12蛋白与靶向合成hpv16dsdna的grna孵育,所述合成hpv16dsdna稀释于人血浆中,终浓度10-18m。误差条表示s.e.m.,n=3。
图5.aacas12b介导的dna检测的特异性和灵敏度。(a)比较没有预扩增的使用ssdna或dsdna激活物的aacas12b反式切割活性。aacas12b与靶向合成ssdna或dsdna的sgrna一起孵育。误差条表示s.e.m.,n=3。(b)aacas12b检测到camvdna的存在。(c)aacas12b区分两种密切相关的合成hpv序列,其具有六个核苷酸多态性。误差棒表示s.e.m.,n=3。(d)比较aacas12b、prcas12a、lbcas12a在区分dsdna方面的特异性,使用合成的hpv18激活物。误差条表示s.e.m.,n=3。(e)使用dsdna激活物比较aacas12b的反式切割活性和预扩增增强的反式切割活性(cdetection)。将aacas12b与靶向靶2的合成ssdna或dsdna的sgrna一起温育。误差条表示s.e.m.,n=3。
图6.cdetection在dna检测中达到亚渺摩尔级灵敏度。(a)在检测和背景基因组混合的hpv16或hpv18dsdna时,cdetection实现亚渺摩尔级
图7.cdetection的应用。(a)通过cdetection检测abo血液基因分型的示意图。示出六种常见abo等位基因和三种靶向性sgrna。各sgrna以可检测信号区分出各自对应的等位基因。如果所有sgrna均没有产生信号,则等位基因为a101或a201。(b)使用cdetection进行abo血液基因分型获得的荧光信号。cdetection未能区分仅有一个单碱基多态性不同的两种dsdna激活物(on-b101vs.off-b101,on-o01vs.off-o01,on-o02/03vs.off-o02/03)。误差棒表示平均值的标准误差(s.e.m.),n=3。rfu,相对荧光单位。(c)示意通过引入经调节的grna(tgrna)开发ecdetection。使用sgrna的cdetection无法区分两种仅有单一错配的dsdna激活物,而使用tgrna(其在间隔区携带错配)的ecdetecion可以通过单核苷酸分辨率实现dna检测。
图8.增强型cdetection(ecdetection)的广泛应用。(a)使用tgrna的ecdection以单核苷酸分辨率特异性实现abo血液基因分型检测。误差棒表示s.e.m.,n=3。rfu,相对荧光单位。tgrna,经调节的grna。(b)和(c)上图示意brca1基因和靶向sgrna以及tgrna之间的序列差异。下图用最大荧光信号示出无rpa的cdetection使用sgrna和tgrna检测人brca1基因突变3232a>g(b)和3537a>g(c)的特异性。(d)荧光时间曲线显示有rpa的cdetection使用tgrna(3232-1)检测人brca1基因突变3232a>g的灵敏度和特异性。野生型brca1或brca13232a>gdsdna稀释于人血浆中,终浓度为10-18m。误差棒表示s.e.m.,n=3。
图9.使用cdetection平台进行精确dna检测。(a)上图示出tp53基因和靶向sgrna以及tgrna之间的序列差异。下图用最大荧光信号示出使用sgrna和tgrna检测人tp53基因突变856g>a的特异性。误差棒表示s.e.m.,n=3。rfu,相对荧光单位。tgrna,经调节的grna。(b)和(c)上图示意brca1基因和靶向sgrna以及tgrna之间的序列差异。下图用最大荧光信号示出使用sgrna和tgrna检测人brca1基因突变3232a>g(b)和3537a>g(c)的特异性。(d)荧光时间曲线显示有rpa的cdetection使用tgrna(3232-1)检测人brca1基因突变3232a>g的灵敏度和特异性。野生型brca1或brca13232a>gdsdna稀释于人血浆中,终浓度为10-16m。误差棒表示s.e.m.,n=3。
图10.cdetection平台的快速及精确的广泛诊断应用。首先通过直接裂解从不同样品获得基因组dna用于临床诊断和定量。对于不同的目的,经过或未经rpa预扩增的dna通过使用sgrna或tgrna的cdetection进行检测。
图11.选择用于基因组编辑测试的非冗余c2c1直系同源物的系统发生树及其基因座。(a)邻接系统发生树,显示测试的c2c1直系同源物的进化关系。(b)对应于(a)中突出显示的8种c2c1蛋白的细菌基因座图谱crrnadr和推定的tracrrna的模拟共折叠显示出稳定的二级结构。dr,直接重复。每个细菌基因组间隔区(spacer)的数目在其crispr阵列的上方或下方表示。
图12.c2c1直系同源物的蛋白质比对:测试的10种c2c1直系同源物的氨基酸序列的多序列比对。保守的残基用红色背景突出显示,保守突变用轮廓和红色字体突出显示。
图13.人293t细胞中c2c1直系同源物介导的基因组靶向。(a)t7ei测定结果表明在人类基因组中与其同源sgrna结合的八种c2c1蛋白的基因组靶向活性。三角形表示切割的条带。(b)t7ei测定结果表明在人293t细胞中由与其同源sgrna(bs3sgrna)结合的bs3c2c1介导的同时多重基因组靶向。(c)sanger测序显示由与bs3sgrna结合的bs3c2c1诱导的代表性插入缺失(indel)。pam和原间隔区序列分别用红色和蓝色着色。插入缺失和插入分别用紫色破折号和绿色小写字符表示。
图14.用于rna指导的基因组编辑的c2c1蛋白。(a)本发明中测试的10种c2c1直系同源物的图形概述。示出其大小(氨基酸数目)。(b)t7ei测定结果表明在人293t细胞中由其同源sgrna指导的八种c2c1直系同源物的基因组靶向活性。三角形表示切割的条带。(c-d)t7ei测定结果表明在人293t细胞中由aasgrna(c)和aksgrna(d)指导的八种c2c1直系同源物的基因组靶向活性。三角形表示切割的条带。
图15.c2c1的sgrna的dna比对:测试衍生自10个c2c1基因座的8种sgrna的dna序列的多序列比对。
图16.不同c2c1直系同源物与sgrna之间的可互换性。t7ei测定结果表明在人293t细胞中由aasgrna(a)、aksgrna(b)、amsgrna(c)、bs3sgrna(d)和lssgrna(e)指导的八种c2c1直系同源物的基因组靶向活性。红色三角形表示切割的条带。
图17.人工sgrna介导的多重基因组靶向。(a)对应于dic2c1和tcc2c1的细菌基因座的图谱。两个c2c1基因座没有crispr阵列。(b-c)t7ei测定结果表明在人293t细胞中由aasgrna(b)和aksgrna(c)指导的aac2c1、dic2c1和tcc2c1的基因组靶向活性。三角形表示切割的条带。(d)t7ei测定结果表明在人293t细胞中由与aksgrna结合的tcc2c1介导的同时多重基因组靶向。(e)示意图说明人工sgrna支架13(artgrna13)的二级结构。(f)t7ei测定结果表明在人293t细胞中由与artgrna13结合的tcc2c1介导的同时多重基因组靶向。
图18.不同sgrna指导c2c1进行基因组编辑。t7ei测定结果表明在人293t细胞中由aasgrna(a)、aksgrna(b)、amsgrna(c)、bs3sgrna(d)和lssgrna(e)指导的aac2c1、dic2c1和tcc2c1的基因组靶向活性。三角形表示切割的条带。
图19.tcc2c1介导的多重基因组编辑。(a)t7ei测定结果表明在人293t细胞中由与amsgrna结合的tcc2c1介导的同时多重基因组靶向。(b-c)sanger测序显示由与aksgrna(b)和amsgrna(c)结合的tcc2c1诱导的代表性插入缺失。pam和原间隔区序列分别用红色和蓝色着色。插入缺失和插入分别用紫色破折号和绿色小写字符表示。
图20.人工sgrna指导tcc2c1进行基因组编辑。(a)示意图说明36种人工sgrna(artgrna)支架(支架:1-12和14-37)的二级结构。(b)t7ei测定结果表明在人293t细胞中artsgrna指导的tcc2c1的基因组靶向活性。三角形表示切割的条带。(c)t7ei测定结果表明在人293t细胞中由与artgrna13结合的aac2c1介导的同时多重基因组靶向。
发明详述
在本发明中,除非另有说明,否则本文中使用的科学和技术名词具有本领域技术人员所通常理解的含义。并且,本文中所用的蛋白质和核酸化学、分子生物学、细胞和组织培养、微生物学、免疫学相关术语和实验室操作步骤均为相应领域内广泛使用的术语和常规步骤。例如,本发明中使用的标准重组dna和分子克隆技术为本领域技术人员熟知,并且在如下文献中有更全面的描述:sambrook,j.,fritsch,e.f.和maniatis,t.,molecularcloning:alaboratorymanual;coldspringharborlaboratorypress:coldspringharbor,1989(下文称为“sambrook”)。同时,为了更好地理解本发明,下面提供相关术语的定义和解释。
在第一方面,本发明提供一种检测生物样品中靶核酸分子的存在和/或量的方法,所述方法包括以下步骤:
(a)使所述生物样品接触:i)cas12b蛋白,ii)针对所述靶核酸分子中的靶序列的grna,和iii)被切割后产生可检测信号的单链dna报告分子,从而形成反应混合物;
(b)检测所述反应混合物中产生的可检测信号的存在和/或水平,
其中所述可检测信号的存在和/或水平代表所述靶核酸分子的存在和/或量。
在一些实施方案中,所述靶核酸分子是双链dna分子。在一些实施方案中,所述靶核酸分子是单链dna分子。所述靶核酸分子可以是基因组dna、cdna、病毒dna等,或它们的片段。
“cas12b”、“cas12b核酸酶”、“cas12b蛋白”、“c2c1”、“c2c1核酸酶”和“c2c1蛋白”在本文中可互换使用,指的是一种来自微生物crispr系统的rna指导的序列特异性核酸酶。cas12b能在向导rna的指导下靶向并切割dna靶序列形成dna双链断裂(dsb),也称作经典dsdna切割活性。更重要的是,cas12b与grna的复合物在识别并结合相应靶dna序列后,能够激活其非特异性单链dna切割活性,也称作非经典旁路ssdna切割活性。利用非经典旁路ssdna切割活性,切割在被切割后产生可检测信号的单链dna报告分子,即可以反映靶dna的存在和/或量。在本文中,被cas12b与grna的复合物识别并结合,激活cas12b的非特异性单链dna切割活性的dna分子也称作“激活物”。
在一些实施方案中,所述cas12b蛋白是来自alicyclobacillusacidiphilus的aacas12b蛋白、来自alicyclobacilluskakegawensis的akcas12b蛋白、来自alicyclobacillusmacrosporangiidus的amcas12b蛋白、来自bacillushisashii的bhcas12b蛋白、来自bacillus属的bscas12b蛋白、来自bacillus属的bs3cas12b蛋白、来自desulfovibrioinopinatus的dicas12b蛋白、来自laceyellasediminis的lscas12b蛋白、来自spirochaetesbacterium的sbcas12b蛋白、来自tuberibacilluscalidus的tccas12b蛋白。在一些优选实施方案中,所述cas12b蛋白是来自alicyclobacillusacidiphilus的cas12b蛋白(aacas12b)。本申请人已经鉴定这些cas12b蛋白可以用于在哺乳动物中进行基因组编辑,也可以用于本发明的核酸检测方法。
例如,所述cas12b蛋白是来自alicyclobacillusacidiphilusnbrc100859的aacas12b蛋白、来自alicyclobacilluskakegawensisnbrc103104的akcas12b蛋白、来自alicyclobacillusmacrosporangiidusstraindsm17980的amcas12b蛋白、来自bacillushisashiistrainc4的bhcas12b蛋白、来自bacillus属nsp2.1的bscas12b蛋白、来自bacillus属v3-13contig_40的bs3cas12b蛋白、来自desulfovibrioinopinatusdsm10711的dicas12b蛋白、来自laceyellasediminisstrainrha1的lscas12b蛋白、来自spirochaetesbacteriumgwb1_27_13的sbcas12b蛋白、来自tuberibacilluscalidusdsm17572的tccas12b蛋白。在一些优选实施方案中,所述cas12b蛋白是来自alicyclobacillusacidiphilusnbrc100859的cas12b蛋白。
alicyclobacillusacidiphilus的cas12b的基因座上缺少已经测序的直接重复(dr)阵列,因此本领域技术人员将会认为其无法进行基因编辑,从而在crispr核酸酶筛选中将其略过。然而,本发明人令人惊奇地发现,来自alicyclobacillusacidiphilus的cas12b蛋白同样具有经典靶向dsdna切割活性和非经典旁路ssdna切割活性,从而能用于基因编辑和核酸检测。类似地,所鉴定的其它一些cas12b蛋白,例如dicas12b或tccas12b蛋白,尽管其天然基因座不具有crispr阵列,也出乎意料地可以用于本发明。
在本发明一些实施方式中,所述cas12b蛋白是其天然基因座不具有crispr阵列的cas12b蛋白。在一些实施方式中,所述天然基因座不具有crispr阵列的cas12b蛋白是aacas12b蛋白、dicas12b或tccas12b蛋白。
在一些实施方案中,所述cas12b蛋白包含与seqidno:1-10中任一个具有至少80%、至少85%、至少90%、至少95%、至少96%、至少97%、至少98%、至少99%序列相同性的氨基酸序列。在一些实施方案中,所述cas12b蛋白的氨基酸序列相对于seqidno:1-10中任一个具有一或多个氨基酸残基取代、缺失或添加。例如,所述cas12b蛋白相对于seqidno:1-10中任一个具有1个、2个、3个、4个、5个、6个、7个、8个、9个或10个氨基酸残基取代、缺失或添加的氨基酸序列。在一些实施方案中,所述氨基酸取代是保守型取代。在一些实施方案中,所述cas12b蛋白包含seqidno:1-10中任一所示的氨基酸序列。例如,所述aacas12b、akcas12b、amcas12b、bhcas12b、bscas12b、bs3cas12b、dicas12b、lscas12b、sbcas12b、tccas12b蛋白分别包含seqidno:1-10所示氨基酸序列。在一些优选实施方式中,所述cas12b蛋白包含seqidno:1所示的氨基酸序列。
本发明人已经证明了cas12b蛋白的ruvc结构域对于其非经典旁路ssdna切割活性是关键的。在一些实施方案中,所述cas12b蛋白包含野生型cas12b蛋白的ruvc结构域,所述野生型cas12b蛋白例如包含seqidno:1-10任一项所示氨基酸序列。本领域技术人员可以容易地鉴定出cas12b蛋白的ruvc结构域。例如,通过ncbi提供的工具。
序列“相同性”具有本领域公认的含义,并且可以利用公开的技术计算两个核酸或多肽分子或区域之间序列相同性的百分比。可以沿着多核苷酸或多肽的全长或者沿着该分子的区域测量序列相同性。(参见,例如:computationalmolecularbiology,lesk,a.m.,ed.,oxforduniversitypress,newyork,1988;biocomputing:informaticsandgenomeprojects,smith,d.w.,ed.,academicpress,newyork,1993;computeranalysisofsequencedata,parti,griffin,a.m.,andgriffin,h.g.,eds.,humanapress,newjersey,1994;sequenceanalysisinmolecularbiology,vonheinje,g.,academicpress,1987;andsequenceanalysisprimer,gribskov,m.anddevereux,j.,eds.,mstocktonpress,newyork,1991)。虽然存在许多测量两个多核苷酸或多肽之间的相同性的方法,但是术语“相同性”是技术人员公知的(carrillo,h.&lipman,d.,siamjappliedmath48:1073(1988))。
在肽或蛋白中,合适的保守型氨基酸取代是本领域技术人员已知的,并且一般可以进行而不改变所得分子的生物活性。通常,本领域技术人员认识到多肽的非必需区中的单个氨基酸取代基本上不改变生物活性(参见,例如,watsonetal.,molecularbiologyofthegene,4thedition,1987,thebenjamin/cummingspub.co.,p.224)。
特别地,本领域人员将可以理解,在同一细菌物种的不同菌株cas12b蛋白可能在氨基酸序列存在一定差异,但是却能实现基本上相同的功能。
在一些实施方案中,所述cas12b蛋白是重组产生的。在一些实施方案中,所述cas12b蛋白还含有融合标签,例如用于cas12b蛋白分离/和或纯化的标签。重组产生蛋白质的方法是本领域已知的。并且本领域已知多种可以用于分离/和或纯化蛋白质的标签,包括但不限于his标签、gst标签等。通常而言,这些标签不会改变目的蛋白的活性。
向导rna”和“grna”在本文中可互换使用,通常由部分互补形成复合物的crrna和tracrrna分子构成,其中crrna包含与靶序列具有足够相同性以便与靶序列的互补序列杂交并且指导crispr复合物(crispr核酸酶+crrna+tracrrna)与该靶序列以序列特异性方式结合的序列。然而,可以设计并使用单向导rna(sgrna),其同时包含crrna和tracrrna的特征。不同的crispr核酸酶对应的grna存在不同。例如,cas9和cas12b通常需要crrna和tracrrna两者,然而,cas12a(cpf1)则只需要crrna。
“针对靶核酸分子的靶序列的grna”指的是grna能够特异性识别所述靶序列。例如,在一些实施方案中(靶核酸分子是双链dna),所述grna包含能够与靶序列的互补序列特异性杂交的间隔区序列(spacer)。在一些实施方案中(靶核酸分子是单链dna),所述grna包含能够与靶序列特异性杂交的间隔区序列。
a.acidiphiluscrispr基因座中没有直接重复序列(dr)阵列,因此,aacas12b并没有对应的crrna。然而,本发明人发现,aacas12b也可以采用来自其他生物体的cas12b蛋白的相应grna。例如aacas12b可以采用自身的tracrrna和来自a.acidoterrestris的crispr基因座的crrna序列作为grna。本发明人对可用于aacas12b的grna进行了优化。
在本发明的一些实施方案中,所述向导rna是由crrna和tracrrna部分互补形成的复合物。在一些实施方案中,所述tracrrna由以下的核酸序列编码:5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaaaaagaacgctcgctcagtgttctgac-3’。在一些实施方案中,所述crrna由以下的核酸序列编码:5’-gtcggatcactgagcgagcgatctgagaagtggcac-nx-3’,其中nx表示x个连续的核苷酸组成的核苷酸序列,n各自独立地选自a、g、c和t;x为18≤x≤35的整数。优选地,x=20。在一些实施方案中,nx是能够与靶序列的互补序列特异性杂交的间隔区序列(靶核酸分子是双链dna)。在一些实施方案中,nx是能够与靶序列特异性杂交的间隔区序列(靶核酸分子是单链dna)。
在本发明的一些实施方案中,所述向导rna是sgrna。在一些实施方案中,所述sgrna由5’端的支架序列和3’端的间隔区序列组成。间隔区序列可以与靶序列或靶序列的互补序列特异性杂交。间隔区序列通常长度为18至35个核苷酸,优选20个核苷酸。
在一些具体实施方案中,所述sgrna由选自以下之一的核酸序列编码:
5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaaaaagaacgctcgctcagtgttctgacgtcggatcactgagcgagcgatctgagaagtggcac-nx-3’;
5’-aactgtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaaaaagaacgctcgctcagtgttctgacgtcggatcactgagcgagcgatctgagaagtggcac-nx-3’;
5’-ctgtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaaaaagaacgctcgctcagtgttctgacgtcggatcactgagcgagcgatctgagaagtggcac-nx-3’;
5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaaaaagaacgctcgctcagtgttatcactgagcgagcgatctgagaagtggcac-nx-3’;
5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaaaaagaacgatctgagaagtggcac-nx-3’;
5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaaaaagctgagaagtggcac-nx-3’;
5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaaagctgagaagtggcac-nx-3’;
5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaaaactgagaagtggcac-nx-3’;
5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaagcgagaagtggcac-nx-3’;
5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctaagcagaagtggcac-nx-3’;和
5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttcaagcgaagtggcac-nx-3’;
其中nx表示x个连续的核苷酸组成的核苷酸序列,n各自独立地选自a、g、c和t;x为18≤x≤35的整数。优选地,x=20。在一些实施方案中,nx是能够与靶序列的互补序列特异性杂交的间隔区序列(靶核酸分子是双链dna)。在一些实施方案中,nx是能够与靶序列特异性杂交的间隔区序列(靶核酸分子是单链dna)。在一些实施方案中,所述sgrna包含由seqidno:11-21中任一项的核苷酸序列编码的支架序列。
在一些具体实施方案中,所述sgrna由选自以下之一的核酸序列编码:
5’-gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttctcaaaaagaacgctcgctcagtgttctgacgtcggatcactgagcgagcgatctgagaagtggcac-nx-3’(aasgrna);
5’-tcgtctataggacggcgaggacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttggcttcaagatgaccgctcgctcagcgatctgacaacggatcgctgagcgagcggtctgagaagtggcac-nx-3’(aksgrna1);
5’-ggaattgccgatctataggacggcagattcaacgggatgtgccaatgcactctttccaggagtgaacaccccgttggcttcaacatgatcgcccgctcaacggtccgatgtcggatcgttgagcgggcgatctgagaagtggcac-nx-3’(amsgrna1);
5’-gaggttctgtcttttggtcaggacaaccgtctagctataagtgctgcagggtgtgagaaactcctattgctggacgatgtctcttttatttcttttttcttggatgtccaagaaaaaagaaatgatacgaggcattagcac-nx-3’(bhsgrna);
5’-ccataagtcgacttacatatccgtgcgtgtgcattatgggcccatccacaggtctattcccacggataatcacgactttccactaagctttcgaatgttcgaaagcttagtggaaagcttcgtggttagcac-nx-3’(bssgrna);
5’-ggtgacctatagggtcaatgaatctgtgcgtgtgccataagtaattaaaaattacccaccacaggattatcttatttctgctaagtgtttagttgcctgaatacttagcagaaataatgatgattggcac-nx-3’(bs3sgrna);
5’-ggcaaagaatactgtgcgtgtgctaaggatggaaaaaatccattcaaccacaggattacattatttatctaatcacttaaatctttaagtgattagatgaattaaatgtgattagcac-nx-3’(lssgrna);或
5’-gtcttagggtatatcccaaatttgtcttagtatgtgcattgcttacagcgacaactaaggtttgtttatcttttttttacattgtaagatgttttacattataaaaagaagataatcttattgcac-nx-3’(sbsgrna);
其中nx表示x个连续的核苷酸组成的核苷酸序列(spacer序列),n各自独立地选自a、g、c和t;x为18≤x≤35的整数。优选地,x=20。在一些实施方案中,序列nx(spacer序列)能够与靶序列的互补序列特异性杂交。所述sgrna中除nx之外的序列为sgrna的支架(scaffold)序列。在一些实施方案中,所述sgrna包含由seqidno:22-29中任一项的核苷酸序列编码的支架序列。
本发明令人惊奇地发现,不同的cas12b系统中的cas12b蛋白以及向导rna可以互换使用,从而使得可以人工设计通用的向导rna。
因此在一些实施方案中,所述sgrna是人工sgrna,其由选自以下的核苷酸序列编码:
5’-ggtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttcaagcgaagtggcac-nx-3’(artsgrna1);
5’-ggtctaaaggacagaagacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttgacttcaagcgaagtggcac-nx-3’(artsgrna2);
5’-ggtctaaaggacagaaaatctgtgcgtgtgccataagtaattaaaaattacccaccacagacttcaagcgaagtggcac-nx-3’(artsgrna3);
5’-ggtcgtctataggacggcgagtttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttcaagcgaagtggcac-nx-3’(artsgrna4);
5’-ggtcgtctataggacggcgaggacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttgacttcaagcgaagtggcac-nx-3’(artsgrna5);
5’-ggtcgtctataggacggcgagaatctgtgcgtgtgccataagtaattaaaaattacccaccacagacttcaagcgaagtggcac-nx-3’(artsgrna6);
5’-ggtgacctatagggtcaatgtttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttcaagcgaagtggcac-nx-3’(artsgrna7);
5’-ggtgacctatagggtcaatggacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttgacttcaagcgaagtggcac-nx-3’(artsgrna8);
5’-ggtgacctatagggtcaatgaatctgtgcgtgtgccataagtaattaaaaattacccaccacagacttcaagcgaagtggcac-nx-3’(artsgrna9);
5’-ggtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgagcttcaaagaagtggcac-nx-3’(artsgrna10);
5’-ggtctaaaggacagaagacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttggcttcaaagaagtggcac-nx-3’(artsgrna11);
5’-ggtctaaaggacagaaaatctgtgcgtgtgccataagtaattaaaaattacccaccacaggcttcaaagaagtggcac-nx-3’(artsgrna12);
5’-ggtcgtctataggacggcgagtttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgagcttcaaagaagtggcac-nx-3’(artsgrna13);
5’-ggtcgtctataggacggcgaggacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttggcttcaaagaagtggcac-nx-3’(artsgrna14);
5’-ggtcgtctataggacggcgagaatctgtgcgtgtgccataagtaattaaaaattacccaccacaggcttcaaagaagtggcac-nx-3’(artsgrna15);
5’-ggtgacctatagggtcaatgtttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgagcttcaaagaagtggcac-nx-3’(artsgrna16);
5’-ggtgacctatagggtcaatggacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttggcttcaaagaagtggcac-nx-3’(artsgrna17);
5’-ggtgacctatagggtcaatgaatctgtgcgtgtgccataagtaattaaaaattacccaccacaggcttcaaagaagtggcac-nx-3’(artsgrna18);
5’-ggtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgagattatctatgatgattggcac-nx-3’(artsgrna19);
5’-ggtctaaaggacagaagacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttggattatctatgatgattggcac-nx-3’(artsgrna20);
5’-ggtctaaaggacagaaaatctgtgcgtgtgccataagtaattaaaaattacccaccacaggattatctatgatgattggcac-nx-3’(artsgrna21);
5’-ggtcgtctataggacggcgagtttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgagattatctatgatgattggcac-nx-3’(artsgrna22);
5’-ggtcgtctataggacggcgaggacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttggattatctatgatgattggcac-nx-3’(artsgrna23);
5’-ggtcgtctataggacggcgagaatctgtgcgtgtgccataagtaattaaaaattacccaccacaggattatctatgatgattggcac-nx-3’(artsgrna24);
5’-ggtgacctatagggtcaatgtttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgagattatctatgatgattggcac-nx-3’(artsgrna25);
5’-ggtgacctatagggtcaatggacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttggattatctatgatgattggcac-nx-3’(artsgrna26);
5’-ggtgacctatagggtcaatgaatctgtgcgtgtgccataagtaattaaaaattacccaccacaggattatctatgatgattggcac-nx-3’(artsgrna27);
5’-ggtctaaaggacagaacaacgggatgtgccaatgcactctttccaggagtgaacaccccgttgacttcaagcgaagtggcac-nx-3’(artsgrna28);
5’-ggtcgtctataggacggcgagcaacgggatgtgccaatgcactctttccaggagtgaacaccccgttgacttcaagcgaagtggcac-nx-3’(artsgrna29);
5’-ggaattgccgatctataggacggcagatttttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgaacttcaagcgaagtggcac-nx-3’(artsgrna30);
5’-ggaattgccgatctataggacggcagattgacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttgacttcaagcgaagtggcac-nx-3’(artsgrna31);
5’-ggaattgccgatctataggacggcagattcaacgggatgtgccaatgcactctttccaggagtgaacaccccgttgacttcaagcgaagtggcac-nx-3’(artsgrna32);
5’-ggtctaaaggacagaacaacgggatgtgccaatgcactctttccaggagtgaacaccccgttggcttcaaagaagtggcac-nx-3’(artsgrna33);
5’-ggtcgtctataggacggcgagcaacgggatgtgccaatgcactctttccaggagtgaacaccccgttggcttcaaagaagtggcac-nx-3’(artsgrna34);
5’-ggaattgccgatctataggacggcagatttttttcaacgggtgtgccaatggccactttccaggtggcaaagcccgttgagcttcaaagaagtggcac-nx-3’(artsgrna35);
5’-ggaattgccgatctataggacggcagattgacaacgggaagtgccaatgtgctctttccaagagcaaacaccccgttggcttcaaagaagtggcac-nx-3’(artsgrn36a);或
5’-ggaattgccgatctataggacggcagattcaacgggatgtgccaatgcactctttccaggagtgaacaccccgttggcttcaaagaagtggcac-nx-3’(artsgrna37),
其中nx表示x个连续的核苷酸组成的核苷酸序列(spacer序列),n各自独立地选自a、g、c和t;x为18≤x≤35的整数。优选地,x=20。在一些实施方案中,序列nx(spacer序列)能够与靶序列的互补序列特异性杂交。所述sgrna中除nx之外的序列为sgrna的支架(scaffold)序列。
在一些实施方案中,所述人工sgrna包含由seqidno:30-66中任一项的核苷酸序列编码的支架序列。
在一些实施方案中,所述grna的间隔区序列被设计为与靶序列或其互补序列完全匹配。在一些实施方案中,所述grna的间隔区序列被设计为与靶序列或其互补序列具有至少一个核苷酸错配,例如具有一个核苷酸错配。这样的grna也称为经调节的(tuned)grna,所述核苷酸错配也成为调节位点。利用cas12b对sgrna与靶之间不同错配的耐受能力差异,设计为与靶序列或其互补序列具有核苷酸错配的grna能够区分靶序列中的单核苷酸多态性变异。在一些实施方案中,所述至少一个核苷酸错配的位置不同于所述单核苷酸多态性变异的位置。例如,经调节的sgrna与靶序列1在位置1具有一个核苷酸错配,而靶序列1与靶序列2在位置2具有单核苷酸多肽性,也即经调节的sgrna与靶序列2之间具有两个核苷酸错配。由于cas12b对错配数目的耐受性差异,其仅在靶序列1存在时产生可检测信号(只有1个错配),而靶序列2没有可检测信号(由于存在两个错配),从而可以将包含单核苷酸多态性的靶序列1和靶序列2区分开。本领域技术人员可以根据具体的靶序列筛选合适的调节位点。
本发明中,grna中除间隔区序列之外的序列也称做grna支架(scaffold)。
在一些实施方案中,所述grna体外转录产生。在一些实施方案中,所述grna通过化学合成产生。
在本发明的一些实施方案中,所述靶核酸分子包含特征性的长度为18-35个核苷酸,优选20个核苷酸的靶序列。在本发明的一些实施方案中,特别是涉及双链dna检测,所述靶序列在紧接其5’端具有选自:5’tttn-3’、5’attn-3’、5’gttn-3’、5’cttn-3’、5’ttc-3’、5’ttg-3’、5’tta-3’、5’ttt-3’、5’tan-3’、5’tgn-3’、5’tcn-3’和5’atc-3’的pam(前间区邻近基序)序列,优选5’tttn-3’。
“被切割后产生可检测信号的单链dna报告分子”例如可以在单链dna的两端分别包含荧光团和其猝灭基团。当单链dna未被切割时,由于猝灭基团存在,荧光团不发出荧光。而当cas12b-grna复合物被靶核酸分子激活,通过其非经典旁路ssdna切割活性切割所述单链dna报告分子的dna单链时,荧光团被释放,从而发出荧光。合适的荧光团及其相应猝灭基团,以及其标记核酸分子的方法在本领域是已知的。合适的荧光团包括但不限于fam、tex、hex、cy3或cy5。合适的猝灭基团包括但不限于bhq1、bhq2、bhq3或tamra。合适的荧光团-猝灭基团对包括但不限于fam-bhq1、tex-bhq2、cy5-bhq3、cy3-bhq1或fam-tamra。因此,在一些实施方案中,所述可检测信号是荧光信号。在一些实施方式中,所述荧光团是fam,所述猝灭基团是bhq1。
所述单链dna报告分子中单链dna的长度可以是大约2-100个核苷酸,例如2-5个、2-10个、2-15个、2-20个、2-25个、2-30个、2-40个或2至更多个核苷酸。所述单链dna报告分子中单链dna可以包含任意序列,但在一些实施方案中,polyg(聚鸟苷酸)除外。在一些实施方案中,所述单链dna报告分子中的单链dna可以选自polya(聚腺苷酸)、polyc(聚胞苷酸)或polyt(聚胸苷酸)。
在一些具体实施方式中,所述单链dna报告分子选自5’-fam-aaaaa-bhq1-3’、5’-fam-ttttt-bhq1-3’、和5’-fam-ccccc-bhq1-3’。
在本发明的方法的一些实施方案中,还包括在步骤(a)之前对所述生物样品中的核酸分子进行扩增的步骤。所述扩增包括但不限于pcr扩增或重组酶聚合酶扩增(recombinasepolymeraseamplification,rpa)。优选地,所述扩增是重组酶聚合酶扩增。
在一些实施方案中,所述重组酶聚合酶扩增进行约10分钟-约60分钟
在一些实施方案中,在与所述生物样品接触之前,所述cas12b蛋白已经与所述grna预先复合形成cas12b-grna复合物。
在一些实施方案中,所述步骤(a)的反应进行约20分钟-约180分钟,例如约20分钟、约30分钟、约40分钟、约50分钟、约60分钟、约90分钟、约120分钟,或之间的任何时间。
在一些实施方案中,步骤(a)在合适的缓冲液中进行。例如,所述缓冲液是nebuffertm2、nebuffertm2.1或
可用于本发明的方法的生物样品包括但不限于全血、血浆、血清、脑脊液、尿液、粪便、细胞或组织提取物等。所述生物样品涵盖提取自细胞或组织的核酸样品。
本发明的范围内还包括用于本发明的方法的试剂盒,该试剂盒包括用于实施本发明的方法的试剂,以及使用说明。例如,所述试剂盒可以包括cas12b蛋白(例如本发明的cas12b蛋白)、grna(例如包含本发明的grna支架)或用于产生grna(例如其包含本发明的grna支架)的试剂、单链dna报告分子(例如本发明的单链dna报告分子),合适的缓冲液、和/或核酸扩增试剂。试剂盒一般包括表明试剂盒内容物的预期用途和/或使用方法的标签。术语标签包括在试剂盒上或与试剂盒一起提供的或以其他方式随试剂盒提供的任何书面的或记录的材料。
本发明还提供了上文所定义的cas12b蛋白和/或包含本发明的支架的grna和/或用于产生包含本发明的支架的grna的试剂在制备用于本发明的方法的试剂盒中的用途。
实施例
实验材料和方法
蛋白表达和纯化
spcas9和lbcas12a蛋白商购获得(neb)。根据之前的报道纯化aacas12b、arcas12a、hkcas12a和prcas12a蛋白。简言之,bpk2014-cas12-his10蛋白在大肠杆菌菌株bl21(λde3)中表达,用0.5mmiptg在16℃诱导表达16小时。收获细胞沉淀并裂解,使用his60nisuperflowresin(takara)纯化。将纯化的cas12蛋白透析,浓缩,最后使用bca蛋白质测定试剂盒(thermofisher)定量。
核酸制备
dna寡核苷酸商购获得(genscript)。双链dna激活物通过pcr扩增获得并使用oligoclean&concentratorkit(zymoresearch)纯化。向导rna使用hiscribetmt7highyieldrnasynthesiskit(neb)体外转录并用microeluternacleanupkit(omega)纯化。
在指定反应中使用的背景基因组dna使用mousedirectpcrkit(bimake)纯化自人胚肾293t细胞。为了模拟无细胞dna(cfdna),dsdna以指定浓度稀释至人血浆中(thermofisher)。
荧光团猝灭剂(fq)-标记的报告测定
用30nmcas12、36nmgrna、混合在40ng背景基因组dna中(在指定的反应中)的40nm激活物(除非另有说明)、200nm定制合成的均聚物ssdnafq报告基因(表1)和nebuffertm2(除非另有说明)在corning384-孔聚苯乙烯nbs微孔板中进行20μl反应检测测定。将反应物在37℃下孵育以在荧光板读数器(bioteksynergy4)中指示时间,每5分钟测量荧光动力学(λex=485nm;λem=528nm,透射增益=61)。通过sigmaplot软件分析荧光结果。
重组酶聚合酶扩增(rpa)反应
根据制造商的方案,使用twistampbasic(twistdx)进行重组酶聚合酶扩增(rpa)反应。将含有不同dna输入量的50μlrpa反应系统在37℃下孵育10分钟。如上所述,将16μlrpa产物直接转移至20μl检测试验。
实施例1、cas12b的反式ssdna切割活性的表征
源自alicyclobacillusacidiphilusnbrc100859的crispr-cas12b核酸酶(aacas12b,氨基酸序列示于seqidno:1)已被用于哺乳动物基因组编辑,因为其具有经典的dsdna靶向切割能力(图1a)。为了表征cas12b的非经典反式旁路切割活性,使用cas12b、cas12a和cas9分别与其相应的向导rna(grna)组合进行体外ssdna切割测定。结果表明,aacas12b和cas12a(arcas12a、hkcas12a和prcas12a)在靶向m13的sgrna(ot-grna)存在下可诱导单链m13dna噬菌体快速降解,而spcas9则不能(图1b)。同样,aacas12b和cas12a在存在与m13噬菌体基因组没有序列同源性的非靶grna及其互补ssdna“激活物”的情况下也实现了m13降解(图1c)。催化失活的变体(发生r785a和d977a取代)中非经典旁路ssdna切割活性被消除,表明该旁路切割活性是ruvc结构域依赖性的(图2)。这些结果表明aacas12b-sgrna复合物一旦被与向导序列互补的dna触发,就可以获得非特异性ssdna反式切割活性。
实施例2、开发cas12b介导的dna检测系统
为了开发cas12b介导的dna检测系统,首先分析了aacas12b-sgrna复合物对荧光团-猝灭剂(fq)标记的同聚物报告分子的切割偏好。发现aacas12b偏好多聚胸苷酸(ployt)以及多聚腺苷酸(polya)和多聚胞苷酸(polyc),而多聚鸟苷酸(polyg)基本上不被切割(图3a)。同时,可以优化切割效率,其在nebuffertm2表现最佳(图3b)。然后,使用nebuffertm2中的polyt报告分子进行aacas12b介导的切割测定。使用sgrna-互补的ot-ssdna和ot-dsdna(ot表示ontarget,在靶),或sgrna-非互补的nt-ssdna和nt-dsdna(nt表述nontarget,非靶)作为激活物,发现了ot-ssdna和ot-dsdna激活物能够触发aacas12b切割fq-报告分子,尽管ot-dsdna效率较低(图3c)。
接下来使用带有不同错配的ssdna或dsdna激活物测试了反式切割活化的特异性。发现pam序列对于dsdna激活物触发的aacas12b的反式切割活性至关重要,并且对于ssdna不是必须的(图4a、b)。同时,还发现dsdna激活物和sgrna之间的错配会阻碍甚至消除反式切割活性,而ssdna激活物则对错配更耐受(图4a、b)。
然后确定了aacas12b-sgrna-激活物系统的灵敏度,发现没有预扩增,aacas12b在ssdna-激活物输入浓度<1.6nm或dsdna-激活物输入浓度<8nm时不产生可检测信号(图5a)。
由于dsdna激活物具有更高的特异性(图4a、b),将aacas12b-sgrna-dsdna激活系统设计为dna检测平台(cas12b-baseddnadetection,cdetection)。合成了一种花椰菜花叶病毒(camv)的dsdna和两种人乳头瘤病毒(hpv16和hpv18)的dsdna作为在检测反应中的激活物。当激活物的输入浓度≥10nm时,aacas12b-sgrna不仅可以产生可检测的信号(图5b,c),还可以区分两种dsdna病毒hpv16和hpv18(图5c)。
与基于cas12a的dna检测相比,aacas12b在这两个检测位点均展示更高的检测灵敏度,因此cdetection产生更高的信号水平和更低的背景水平(图4c和图5d)。
为了提高灵敏度,使用重组酶聚合酶扩增(rpa)进行了预扩增,并在
实施例3、开发增强型cas12b介导的dna检测系统
为了扩展和模拟cdetection在分子诊断应用中的应用,将合成的hpvdsdna稀释到人类基因组dna中。结果显示cdetection可以在亚渺摩尔级
aacas12b在人血浆中的高灵敏度促使测试cdetection在基于cfdna的非侵入性诊断中的应用。虽然之前使用过cfdna分析灵敏度达到了108分之一,但这些方法需要相对大量的cfdna(5-10ng/ml血浆),并且是耗时的。为了证明cdetection在cfdna检测中的优势,将hpv的dsdna稀释进人血浆中并检测该新方法的灵敏度。结果显示,cdetection可以检测人血浆中浓度低至
为了扩展cdetection在精确诊断中的应用,我们设计了使用三种靶向sgrna和相应的dsdna激活物(on-以及off-激活物)的实验来鉴定六种常见的人abo等位基因。理论上,携带三种sgrna中的每一种的cdetection可分别鉴定出o01、o02/o03和b101。如果所有sgrna的荧光信号没有检测到,则等位基因应为a101/a201(图7a)。结果显示,cdetection无法区分不同的abo等位基因,因为它产生在on和off的dsdna激活物组之间难以区分的荧光信号(图7b)。
为了增强cdetection的特异性,设计了在间隔区序列(spacer)中含有单核苷酸错配的经调节的向导rna(tunedgrna,tgrna),其使得只有单个核苷酸差异的两种相似靶标由不可区分变为可区分(图7c)。
为了证明这种增强的cdetection(ecdetection)的单碱基分辨率灵敏度,进行了abo血液基因分型测试。结果表明,ecdetection可以高精度地确定血型,而cdetection则不能(图8a)。
测序和探针检测是检测点突变引起的疾病的两种主要方法。然而,测序是昂贵且耗时的,并且其灵敏度取决于测序深度。基于探针的方法对单碱基突变的敏感性较差。由于本发明的ecdetection方法具有高特异性和灵敏度,可以使用ecdetection检测人类基因组中的低比率单碱基突变。
选择了癌症相关的tp53856g>a突变来测试其可行性。结果表明,cdetection可以使用选定的tgrna准确区分点突变等位基因与野生型等位基因(图9a)。
此外,应用cdetection平台检测乳腺癌相关brca1基因的两个热点(3232a>g和3537a>g)。用选择的tgrna(tgrna-3232-1和tgrna-3537-4)进行cdetection可以很好地区分点突变,而sgrna几乎不支持点突变检测(图8b、c和图9b、c)。
此外,为了模拟通过cdetection使用cfdna早期临床检测原发疾病,将brca13232a>gdsdna稀释到人血浆中。研究结果表明,cdetection可以在单碱基分辨率下实现点突变检测(图8d和图9d)。本发明的ecdetection能够以单碱基分辨率在临床研究实现快速dna检测。
总之,本发明提供了cdetection平台,该平台基于cas12b核酸酶的非经典旁路ssdna切割特性,可以渺摩尔级灵敏度检测dna。同时,结合经调节的grna,开发了增强版(ecdetection),实现单核苷酸分辨率。本发明的cdetection和ecdetection平台可以在各种分子诊断应用中更容易地检测核酸,以及在临床研究中进行基因型分析(图10)。
实施例4、其他cas12b蛋白的鉴定
选择并从头合成来自不同细菌的六种代表性cas12b蛋白,以及之前报道的四种cas12b直系同源物,在人胚胎肾293t细胞中进行基因组编辑(图11、12和seqidno:1-10)。在这10种cas12b直系同源物中,来自d.inopinatus(dicas12b)和t.calidus(tccas12b)的cas12b既没有可预测的前体crisprrna(pre-crrna)也没有反式激活crrna(tracrrna)(图11b),提示这两种cas12b蛋白可能不适合基因组编辑应用。
为了进行哺乳动物基因组编辑,用单独的cas12b酶和其靶向含有适当pam的人内源基因座的同源单向导rna(sgrna)共转染293t细胞(图11)。t7核酸内切酶(t7ei)测定的结果显示,aacas12b、akcas12b、amcas12b、bhcas12b、bs3cas12b和lscas12b稳健地编辑人类基因组,尽管它们的靶向效率在不同的直系同源物之间和在不同的靶向位点不同(图11b和图13a)。还通过简单地使用多个sgrna,使用bs3cas12b实现多重基因组编辑,同时编辑人类基因组中的四个位点(图13b,c)。这些新发现的有功能的cas12b直系同源物扩展对基于cas12b的基因组编辑的选择,同时扩展了基于cas12b的dna检测的选择。
实施例5、不同cas12b及双rna的可互换性
为了研究cas12b系统中双rna(crrna和tracrrna)和蛋白质组分之间的可互换性,首先分析cas12b蛋白和双rna两者的保守性。除了cas12b直系同源物的保守氨基酸序列外(图14a和图12),前体crrna:tracrrna双链体的dna序列及其二级结构也表现出高保守性(图11b和图15)。接下来,用分别与来自8个cas12b系统的各sgrna复合的8种cas12b直系同源物,在293t细胞中进行基因组编辑。如t7ei测定的结果所示,衍生自aacas12b、akcas12b、amcas12b、bs3cas12b和lscas12b基因座的sgrna可以替代原始sgrna用于哺乳动物基因组编辑,尽管在不同cas12b直系同源物和sgrna之间的活性有所不同(图14c,d和图16)。这些结果证明不同cas12b和来自不同cas12b基因座的双rna之间的可互换性。
实施例6、利用天然基因座无crispr阵列的cas12b进行基因组编辑
本发明进一步选择两个基因座没有携带crispr阵列的cas12b直系同源物dicas12b和tccas12b进行后续实验(图17a)。基因座没有携带crispr阵列使得它们的crrna:tracrrna双链体的序列不可预测。在293t细胞中共转染与靶向不同基因组位点的衍生自其他8种cas12b直系同源物的基因座的sgrna组合的dicas12b和tccas12b以及aacas12b。t7ei测定结果表明衍生自aacas12b、akcas12b、amcas12b、bs3cas12b和lscas12b的sgrna使tccas12b能够稳健地编辑人类基因组(图17b、c和图18)。此外,aasgrna或aksgrna能够使tccas12b实现多重基因组编辑(图17d和图19)。上述结果表明在来自不同系统的cas12b和双链rna之间可互换性使得可能利用天然基因座不具有crispr阵列的cas12b直系同源物来编辑哺乳动物基因组。
实施例7、设计用于cas12b介导的基因组编辑的人工sgrna
不同cas12b系统中cas12b蛋白和双rna之间的可互换性有利于设计新的人工sgrna(artsgrna)支架以促进cas12b介导的基因组编辑。考虑到cas12b直系同源物中dna序列和二级结构的保守性(图11b和13),设计并从头合成37种sgrna支架(seqidno:30-66),用于靶向人ccr5基因座(图17e,图20a)。t7ei测定的结果表明22种artsgrna支架有效地工作(图10b)。为了验证artgrna的普遍适用性,使用artsgrna13指导tccas12b或aacas12b进行多重基因组编辑(图20a)。t7ei测定结果表明,artsgrna13同时促进tccas12b和aacas12b两者的多重基因组编辑(图17f和图20c)。结果表明通过设计和合成artsgrna能促进cas12b介导的基因组编辑特别是多重基因组编辑。
表1.实施例中涉及的核酸序列(其中下划线为spacer序列,粗体斜体为tgrna中的错配核苷酸)
表2.cas12b-介导的dna检测中测试的缓冲液的组分。缓冲液1、3、4、5来自已有的报道,而缓冲液2、6、7、8来自商品化的缓冲液。1:nucleaseassaybuffer;2:nebuffertm3.1;3:cas12abindingbuffer;4:cas13buffer;5:cas12abuffer;6:nebuffertm2;7:nebuffertm2.1;8:
表3.本发明的序列表及对应的序列信息
序列表
<110>中国科学院动物研究所
<120>核酸检测方法
<130>tc2737
<150>201811099146.0
<151>2018-09-20
<160>66
<170>patentinversion3.5
<210>1
<211>1129
<212>prt
<213>alicyclobacillusacidiphilus
<400>1
metalavallyssermetlysvallysleuargleuaspasnmetpro
151015
gluileargalaglyleutrplysleuhisthrgluvalasnalagly
202530
valargtyrtyrthrglutrpleuserleuleuargglngluasnleu
354045
tyrargargserproasnglyaspglygluglnglucystyrlysthr
505560
alagluglucyslysalagluleuleugluargleuargalaarggln
65707580
valgluasnglyhiscysglyproalaglyseraspaspgluleuleu
859095
glnleualaargglnleutyrgluleuleuvalproglnalailegly
100105110
alalysglyaspalaglnglnilealaarglyspheleuserproleu
115120125
alaasplysaspalavalglyglyleuglyilealalysalaglyasn
130135140
lysproargtrpvalargmetargglualaglygluproglytrpglu
145150155160
gluglulysalalysalaglualaarglysserthraspargthrala
165170175
aspvalleuargalaleualaasppheglyleulysproleumetarg
180185190
valtyrthraspseraspmetserservalglntrplysproleuarg
195200205
lysglyglnalavalargthrtrpaspargaspmetpheglnglnala
210215220
ilegluargmetmetsertrpglusertrpasnglnargvalglyglu
225230235240
alatyralalysleuvalgluglnlysserargphegluglnlysasn
245250255
phevalglyglngluhisleuvalglnleuvalasnglnleuglngln
260265270
aspmetlysglualaserhisglyleugluserlysgluglnthrala
275280285
histyrleuthrglyargalaleuargglyserasplysvalpheglu
290295300
lystrpglulysleuaspproaspalapropheaspleutyraspthr
305310315320
gluilelysasnvalglnargargasnthrargargpheglyserhis
325330335
aspleuphealalysleualagluprolystyrglnalaleutrparg
340345350
gluaspalaserpheleuthrargtyralavaltyrasnserileval
355360365
arglysleuasnhisalalysmetphealathrphethrleuproasp
370375380
alathralahisproiletrpthrargpheasplysleuglyglyasn
385390395400
leuhisglntyrthrpheleupheasnglupheglygluglyarghis
405410415
alaileargpheglnlysleuleuthrvalgluaspglyvalalalys
420425430
gluvalaspaspvalthrvalproilesermetseralaglnleuasp
435440445
aspleuleuproargaspprohisgluleuvalalaleutyrphegln
450455460
asptyrglyalagluglnhisleualaglyglupheglyglyalalys
465470475480
ileglntyrargargaspglnleuasnhisleuhisalaargarggly
485490495
alaargaspvaltyrleuasnleuservalargvalglnserglnser
500505510
glualaargglygluargargproprotyralaalavalpheargleu
515520525
valglyaspasnhisargalaphevalhispheasplysleuserasp
530535540
tyrleualagluhisproaspaspglylysleuglysergluglyleu
545550555560
leuserglyleuargvalmetservalaspleuglyleuargthrser
565570575
alaserileservalpheargvalalaarglysaspgluleulyspro
580585590
asnsergluglyargvalprophecyspheproilegluglyasnglu
595600605
asnleuvalalavalhisgluargserglnleuleulysleuprogly
610615620
gluthrgluserlysaspleuargalaileargglugluargglnarg
625630635640
thrleuargglnleuargthrglnleualatyrleuargleuleuval
645650655
argcysglysergluaspvalglyargarggluargsertrpalalys
660665670
leuilegluglnprometaspalaasnglnmetthrproasptrparg
675680685
glualaphegluaspgluleuglnlysleulysserleutyrglyile
690695700
cysglyaspargglutrpthrglualavaltyrgluservalargarg
705710715720
valtrparghismetglylysglnvalargasptrparglysaspval
725730735
argserglygluargprolysileargglytyrglnlysaspvalval
740745750
glyglyasnserilegluglnileglutyrleugluargglntyrlys
755760765
pheleulyssertrpserphepheglylysvalserglyglnvalile
770775780
argalaglulysglyserargphealailethrleuarggluhisile
785790795800
asphisalalysgluaspargleulyslysleualaaspargileile
805810815
metglualaleuglytyrvaltyralaleuaspaspgluargglylys
820825830
glylystrpvalalalystyrproprocysglnleuileleuleuglu
835840845
gluleuserglutyrglnpheasnasnaspargproprosergluasn
850855860
asnglnleumetglntrpserhisargglyvalpheglngluleuleu
865870875880
asnglnalaglnvalhisaspleuleuvalglythrmettyralaala
885890895
pheserserargpheaspalaargthrglyalaproglyileargcys
900905910
argargvalproalaargcysalaarggluglnasnprogluprophe
915920925
protrptrpleuasnlysphevalalagluhislysleuaspglycys
930935940
proleuargalaaspaspleuileprothrglygluglygluphephe
945950955960
valserpropheseralaglugluglyaspphehisglnilehisala
965970975
aspleuasnalaalaglnasnleuglnargargleutrpseraspphe
980985990
aspileserglnileargleuargcysasptrpglygluvalaspgly
99510001005
gluprovalleuileproargthrthrglylysargthralaasp
101010151020
sertyrglyasnlysvalphetyrthrlysthrglyvalthrtyr
102510301035
tyrgluarggluargglylyslysargarglysvalphealagln
104010451050
glugluleuserglugluglualagluleuleuvalglualaasp
105510601065
glualaargglulysservalvalleumetargaspprosergly
107010751080
ileileasnargglyasptrpthrargglnlysgluphetrpser
108510901095
metvalasnglnargilegluglytyrleuvallysglnilearg
110011051110
serargvalargleuglngluseralacysgluasnthrglyasp
111511201125
ile
<210>2
<211>1147
<212>prt
<213>alicyclobacilluskakegawensis
<400>2
metalavallysserilelysvallysleuargleuserglucyspro
151015
aspileleualaglymettrpglnleuhisargalathrasnalagly
202530
valargtyrtyrthrglutrpvalserleumetargglngluileleu
354045
tyrserargglyproaspglyglyglnglncystyrmetthralaglu
505560
aspcysglnarggluleuleuargargleuargasnargglnleuhis
65707580
asnglyargglnaspglnproglythraspalaaspleuleualaile
859095
serargargleutyrgluileleuvalleuglnserileglylysarg
100105110
glyaspalaglnglnilealaserserpheleuserproleuvalasp
115120125
proasnserlysglyglyargglyglualalysserglyarglyspro
130135140
alatrpglnlysmetargaspglnglyaspproargtrpvalalaala
145150155160
argglulystyrgluglnarglysalavalaspproserlysgluile
165170175
leuasnserleuaspalaleuglyleuargproleuphealavalphe
180185190
thrgluthrtyrargserglyvalasptrplysproleuglylysser
195200205
glnglyvalargthrtrpaspargaspmetpheglnglnalaleuglu
210215220
argleumetsertrpglusertrpasnargargvalglygluglutyr
225230235240
alaargleupheglnglnlysmetlysphegluglngluhispheala
245250255
gluglnserhisleuvallysleualaargalaleuglualaaspmet
260265270
argalaalaserglnglypheglualalysargglythralahisgln
275280285
ilethrargargalaleuargglyalaaspargvalphegluiletrp
290295300
lysserileprogluglualaleupheserglntyraspgluvalile
305310315320
argglnvalglnalaglulysargargasppheglyserhisaspleu
325330335
phealalysleualagluprolystyrglnproleutrpargalaasp
340345350
gluthrpheleuthrargtyralaleutyrasnglyvalleuargasp
355360365
leuglulysalaargglnphealathrphethrleuproaspalacys
370375380
valasnproiletrpthrargphegluserserglnglyserasnleu
385390395400
hislystyrglupheleupheasphisleuglyproglyarghisala
405410415
valargpheglnargleuleuvalvalglusergluglyalalysglu
420425430
argaspservalvalvalprovalalaproserglyglnleuasplys
435440445
leuvalleuargglugluglulysserservalalaleuhisleuhis
450455460
aspthralaargproaspglyphemetalaglutrpalaglyalalys
465470475480
leuglntyrgluargserthrleualaarglysalaargargasplys
485490495
glnglymetargsertrpargargglnprosermetleumetserala
500505510
alaglnmetleugluaspalalysglnalaglyaspvaltyrleuasn
515520525
ileservalargvallysserprosergluvalargglyglnargarg
530535540
proprotyralaalaleupheargileaspasplysglnargargval
545550555560
thrvalasntyrasnlysleuseralatyrleuglugluhisproasp
565570575
lysglnileproglyalaproglyleuleuserglyleuargvalmet
580585590
servalaspleuglyleuargthrseralaserileservalphearg
595600605
valalalyslysglugluvalglualaleuglyaspglyargpropro
610615620
histyrtyrproilehisglythraspaspleuvalalavalhisglu
625630635640
argserhisleuileglnmetproglygluthrgluthrlysglnleu
645650655
arglysleuargglugluargglnalavalleuargproleupheala
660665670
glnleualaleuleuargleuleuvalargcysglyalaalaaspglu
675680685
argileargthrargsertrpglnargleuthrlysglnglyargglu
690695700
phethrlysargleuthrprosertrpargglualaleugluleuglu
705710715720
leuthrargleuglualatyrcysglyargvalproaspaspglutrp
725730735
serargilevalaspargthrvalilealaleutrpargargmetgly
740745750
lysglnvalargasptrparglysglnvallysserglyalalysval
755760765
lysvallysglytyrglnleuaspvalvalglyglyasnserleuala
770775780
glnileasptyrleugluglnglntyrlyspheleuargargtrpser
785790795800
phephealaargalaserglyleuvalvalargalaasparggluser
805810815
hisphealavalalaleuargglnhisilegluasnalalysargasp
820825830
argleulyslysleualaaspargileleumetglualaleuglytyr
835840845
valtyrglualaserglyproarggluglyglntrpthralaglnhis
850855860
proprocysglnleuileileleuglugluleuseralatyrargphe
865870875880
seraspaspargproprosergluasnserlysleumetalatrpgly
885890895
hisargglyileleuglugluleuvalasnglnalaglnvalhisasp
900905910
valleuvalglythrvaltyralaalapheserserargpheaspala
915920925
argthrglyalaproglyvalargcysargargvalproalaargphe
930935940
valglyalathrvalaspaspserleuproleutrpleuthrgluphe
945950955960
leuasplyshisargleuasplysasnleuleuargproaspaspval
965970975
ileprothrglygluglyglupheleuvalserprocysglygluglu
980985990
alaalaargvalargglnvalhisalaaspileasnalaalaglnasn
99510001005
leuglnargargleutrpglnasnpheaspilethrgluleuarg
101010151020
leuargcysaspvallysmetglyglygluglythrvalleuval
102510301035
proargvalasnasnalaargalalysglnleupheglylyslys
104010451050
valleuvalserglnaspglyvalthrphephegluargsergln
105510601065
thrglyglylysprohisserglulysglnthraspleuthrasp
107010751080
lysgluleugluleuilealaglualaaspglualaargalalys
108510901095
servalvalleupheargaspproserglyhisileglylysgly
110011051110
histrpileargglnarggluphetrpserleuvallysglnarg
111511201125
ilegluserhisthralagluargileargvalargglyvalgly
113011351140
serserleuasp
1145
<210>3
<211>1146
<212>prt
<213>alicyclobacillusmacrosporangiidus
<400>3
metasnvalalavallysserilelysvallysleumetleuglyhis
151015
leuprogluilearggluglyleutrphisleuhisglualavalasn
202530
leuglyvalargtyrtyrthrglutrpleualaleuleuargglngly
354045
asnleutyrargargglylysaspglyalaglnglucystyrmetthr
505560
alagluglncysargglngluleuleuvalargleuargasparggln
65707580
lysargasnglyhisthrglyaspproglythraspglugluleuleu
859095
glyvalalaargargleutyrgluleuleuvalproglnservalgly
100105110
lyslysglyglnalaglnmetleualaserglypheleuserproleu
115120125
alaaspprolyssergluglyglylysglythrserlysserglyarg
130135140
lysproalatrpmetglymetlysglualaglyaspserargtrpval
145150155160
glualalysalaargtyrglualaasnlysalalysaspprothrlys
165170175
glnvalilealaserleuglumettyrglyleuargproleupheasp
180185190
valphethrgluthrtyrlysthrileargtrpmetproleuglylys
195200205
hisglnglyvalargalatrpaspargaspmetpheglnglnserleu
210215220
gluargleumetsertrpglusertrpasngluargvalglyalaglu
225230235240
phealaargleuvalaspargargaspargpheargglulyshisphe
245250255
thrglyglngluhisleuvalalaleualaglnargleugluglnglu
260265270
metlysglualaserproglyphegluserlysserserglnalahis
275280285
argilethrlysargalaleuargglyalaaspglyileileaspasp
290295300
trpleulysleusergluglygluprovalaspargpheaspgluile
305310315320
leuarglysargglnalaglnasnproargargpheglyserhisasp
325330335
leupheleulysleualagluprovalpheglnproleutrpargglu
340345350
aspproserpheleuserargtrpalasertyrasngluvalleuasn
355360365
lysleugluaspalalysglnphealathrphethrleuproserpro
370375380
cysserasnprovaltrpalaargphegluasnalagluglythrasn
385390395400
ilephelystyrasppheleupheasphispheglylysglyarghis
405410415
glyvalargpheglnargmetilevalmetargaspglyvalprothr
420425430
gluvalgluglyilevalvalproilealaproserargglnleuasp
435440445
alaleualaproasnaspalaalaserproileaspvalphevalgly
450455460
aspproalaalaproglyalapheargglyglnpheglyglyalalys
465470475480
ileglntyrargargseralaleuvalarglysglyargarggluglu
485490495
lysalatyrleucysglypheargleuproserglnargargthrgly
500505510
thrproalaaspaspalaglygluvalpheleuasnleuserleuarg
515520525
valgluserglnsergluglnalaglyargargasnproprotyrala
530535540
alavalphehisileseraspglnthrargargvalilevalargtyr
545550555560
glygluilegluargtyrleualagluhisproaspthrglyilepro
565570575
glyserargglyleuthrserglyleuargvalmetservalaspleu
580585590
glyleuargthrseralaalaileservalpheargvalalahisarg
595600605
aspgluleuthrproaspalahisglyargglnprophephephepro
610615620
ilehisglymetasphisleuvalalaleuhisgluargserhisleu
625630635640
ileargleuproglygluthrgluserlyslysvalargserilearg
645650655
gluglnargleuaspargleuasnargleuargserglnmetalaser
660665670
leuargleuleuvalargthrglyvalleuaspgluglnlysargasp
675680685
argasntrpgluargleuglnsersermetgluargglyglygluarg
690695700
metproserasptrptrpaspleupheglnalaglnvalargtyrleu
705710715720
alaglnhisargaspalaserglyglualatrpglyargmetvalgln
725730735
alaalavalargthrleutrpargglnleualalysglnvalargasp
740745750
trparglysgluvalargargasnalaasplysvallysilearggly
755760765
ilealaargaspvalproglyglyhisserleualaglnleuasptyr
770775780
leugluargglntyrargpheleuargsertrpseralapheserval
785790795800
glnalaglyglnvalvalargalagluargaspserargphealaval
805810815
alaleuarggluhisileaspasnglylyslysaspargleulyslys
820825830
leualaaspargileleumetglualaleuglytyrvaltyrvalthr
835840845
aspglyargargalaglyglntrpglnalavaltyrproprocysgln
850855860
leuvalleuleuglugluleuserglutyrargpheserasnasparg
865870875880
proprosergluasnserglnleumetvaltrpserhisargglyval
885890895
leuglugluleuilehisglnalaglnvalhisaspvalleuvalgly
900905910
thrileproalaalapheserserargpheaspalaargthrglyala
915920925
proglyileargcysargargvalproserileproleulysaspala
930935940
proserileproiletrpleuserhistyrleulysglnthrgluarg
945950955960
aspalaalaalaleuargproglygluleuileprothrglyaspgly
965970975
glupheleuvalthrproalaglyargglyalaserglyvalargval
980985990
valhisalaaspileasnalaalahisasnleuglnargargleutrp
99510001005
gluasnpheaspleuseraspileargvalargcysaspargarg
101010151020
gluglylysaspglythrvalvalleuileproargleuthrasn
102510301035
glnargvallysgluargtyrserglyvalilephethrserglu
104010451050
aspglyvalserphethrvalglyaspalalysthrargargarg
105510601065
serseralaserglnglygluglyaspaspleuseraspgluglu
107010751080
glngluleuleualaglualaaspaspalaarggluargserval
108510901095
valleupheargaspproserglyphevalasnglyglyargtrp
110011051110
thralaglnargalaphetrpglymetvalhisasnargileglu
111511201125
thrleuleualagluargpheservalserglyalaalaglulys
113011351140
valarggly
1145
<210>4
<211>1108
<212>prt
<213>bacillushisashii
<400>4
metalathrargserpheileleulysilegluproasnglugluval
151015
lyslysglyleutrplysthrhisgluvalleuasnhisglyileala
202530
tyrtyrmetasnileleulysleuileargglnglualailetyrglu
354045
hishisgluglnaspprolysasnprolyslysvalserlysalaglu
505560
ileglnalagluleutrpaspphevalleulysmetglnlyscysasn
65707580
serphethrhisgluvalasplysaspgluvalpheasnileleuarg
859095
gluleutyrglugluleuvalproserservalglulyslysglyglu
100105110
alaasnglnleuserasnlyspheleutyrproleuvalaspproasn
115120125
serglnserglylysglythralaserserglyarglysproargtrp
130135140
tyrasnleulysilealaglyaspprosertrpglugluglulyslys
145150155160
lystrpglugluasplyslyslysaspproleualalysileleugly
165170175
lysleualaglutyrglyleuileproleupheileprotyrthrasp
180185190
serasngluproilevallysgluilelystrpmetglulysserarg
195200205
asnglnservalargargleuasplysaspmetpheileglnalaleu
210215220
gluargpheleusertrpglusertrpasnleulysvallysgluglu
225230235240
tyrglulysvalglulysglutyrlysthrleuglugluargilelys
245250255
gluaspileglnalaleulysalaleugluglntyrglulysgluarg
260265270
glngluglnleuleuargaspthrleuasnthrasnglutyrargleu
275280285
serlysargglyleuargglytrparggluileileglnlystrpleu
290295300
lysmetaspgluasngluproserglulystyrleugluvalphelys
305310315320
asptyrglnarglyshisproargglualaglyasptyrservaltyr
325330335
glupheleuserlyslysgluasnhispheiletrpargasnhispro
340345350
glutyrprotyrleutyralathrphecysgluileasplyslyslys
355360365
lysaspalalysglnglnalathrphethrleualaaspproileasn
370375380
hisproleutrpvalargpheglugluargserglyserasnleuasn
385390395400
lystyrargileleuthrgluglnleuhisthrglulysleulyslys
405410415
lysleuthrvalglnleuaspargleuiletyrprothrglusergly
420425430
glytrpgluglulysglylysvalaspilevalleuleuproserarg
435440445
glnphetyrasnglnilepheleuaspilegluglulysglylyshis
450455460
alaphethrtyrlysaspgluserilelyspheproleulysglythr
465470475480
leuglyglyalaargvalglnpheaspargasphisleuargargtyr
485490495
prohislysvalgluserglyasnvalglyargiletyrpheasnmet
500505510
thrvalasnilegluprothrgluserprovalserlysserleulys
515520525
ilehisargaspasppheprolysvalvalasnphelysprolysglu
530535540
leuthrglutrpilelysaspserlysglylyslysleulyssergly
545550555560
ilegluserleugluileglyleuargvalmetserileaspleugly
565570575
glnargglnalaalaalaalaserilephegluvalvalaspglnlys
580585590
proaspilegluglylysleuphepheproilelysglythrgluleu
595600605
tyralavalhisargalaserpheasnilelysleuproglygluthr
610615620
leuvallysserarggluvalleuarglysalaarggluaspasnleu
625630635640
lysleumetasnglnlysleuasnpheleuargasnvalleuhisphe
645650655
glnglnphegluaspilethrgluargglulysargvalthrlystrp
660665670
ileserargglngluasnseraspvalproleuvaltyrglnaspglu
675680685
leuileglnilearggluleumettyrlysprotyrlysasptrpval
690695700
alapheleulysglnleuhislysargleugluvalgluileglylys
705710715720
gluvallyshistrparglysserleuseraspglyarglysglyleu
725730735
tyrglyileserleulysasnileaspgluileaspargthrarglys
740745750
pheleuleuargtrpserleuargprothrgluproglygluvalarg
755760765
argleugluproglyglnargphealaileaspglnleuasnhisleu
770775780
asnalaleulysgluaspargleulyslysmetalaasnthrileile
785790795800
methisalaleuglytyrcystyraspvalarglyslyslystrpgln
805810815
alalysasnproalacysglnileileleuphegluaspleuserasn
820825830
tyrasnprotyrglugluargserargphegluasnserlysleumet
835840845
lystrpserargarggluileproargglnvalalaleuglnglyglu
850855860
iletyrglyleuglnvalglygluvalglyalaglnpheserserarg
865870875880
phehisalalysthrglyserproglyileargcysservalvalthr
885890895
lysglulysleuglnaspasnargphephelysasnleuglnargglu
900905910
glyargleuthrleuasplysilealavalleulysgluglyaspleu
915920925
tyrproasplysglyglyglulyspheileserleuserlysasparg
930935940
lyscysvalthrthrhisalaaspileasnalaalaglnasnleugln
945950955960
lysargphetrpthrargthrhisglyphetyrlysvaltyrcyslys
965970975
alatyrglnvalaspglyglnthrvaltyrileprogluserlysasp
980985990
glnlysglnlysileilegluglupheglygluglytyrpheileleu
99510001005
lysaspglyvaltyrglutrpvalasnalaglylysleulysile
101010151020
lyslysglyserserlysglnsersersergluleuvalaspser
102510301035
aspileleulysaspserpheaspleualasergluleulysgly
104010451050
glulysleumetleutyrargaspproserglyasnvalphepro
105510601065
serasplystrpmetalaalaglyvalphepheglylysleuglu
107010751080
argileleuileserlysleuthrasnglntyrserileserthr
108510901095
ilegluaspaspserserlysglnsermet
11001105
<210>5
<211>1108
<212>prt
<213>bacillus
<400>5
metalaileargserilelysleulysleulysthrhisthrglypro
151015
glualaglnasnleuarglysglyiletrpargthrhisargleuleu
202530
asngluglyvalalatyrtyrmetlysmetleuleuleuphearggln
354045
gluserthrglygluargprolysglugluleuglnglugluleuile
505560
cyshisilearggluglnglnglnargasnglnalaasplysasnthr
65707580
glnalaleuproleuasplysalaleuglualaleuargglnleutyr
859095
gluleuleuvalproserservalglyglnserglyaspalaglnile
100105110
ileserarglyspheleuserproleuvalaspproasnserglugly
115120125
glylysglythrserlysalaglyalalysprothrtrpglnlyslys
130135140
lysglualaasnaspprothrtrpgluglnasptyrglulystrplys
145150155160
lysargargglugluaspprothralaservalilethrthrleuglu
165170175
glutyrglyileargproilepheproleutyrthrasnthrvalthr
180185190
aspilealatrpleuproleuglnserasnglnphevalargthrtrp
195200205
aspargaspmetleuglnglnalailegluargleuleusertrpglu
210215220
sertrpasnlysargvalglngluglutyralalysleulysglulys
225230235240
metalaglnleuasngluglnleugluglyglyglnglutrpileser
245250255
leuleugluglntyrglugluasnarggluarggluleuarggluasn
260265270
metthralaalaasnasplystyrargilethrlysargglnmetlys
275280285
glytrpasngluleutyrgluleutrpserthrpheproalaserala
290295300
serhisgluglntyrlysglualaleulysargvalglnglnargleu
305310315320
argglyargpheglyaspalahisphepheglntyrleumetgluglu
325330335
lysasnargleuiletrplysglyasnproglnargilehistyrphe
340345350
valalaargasngluleuthrlysargleugluglualalysglnser
355360365
alathrmetthrleuproasnalaarglyshisproleutrpvalarg
370375380
pheaspalaargglyglyasnleuglnasptyrtyrleuthralaglu
385390395400
alaasplysproargserargargphevalthrpheserglnleuile
405410415
trpprosergluserglytrpmetglulyslysaspvalgluvalglu
420425430
leualaleuserargglnphetyrglnglnvallysleuleulysasn
435440445
asplysglylysglnlysilegluphelysasplysglyserglyser
450455460
thrpheasnglyhisleuglyglyalalysleuglnleugluarggly
465470475480
aspleuglulysgluglulysasnphegluaspglygluileglyser
485490495
valtyrleuasnvalvalileaspphegluproleuglngluvallys
500505510
asnglyargvalglnalaprotyrglyglnvalleuglnleuilearg
515520525
argproasnglupheprolysvalthrthrtyrlyssergluglnleu
530535540
valglutrpilelysalaserproglnhisseralaglyvalgluser
545550555560
leualaserglypheargvalmetserileaspleuglyleuargala
565570575
alaalaalathrserilepheservalglugluserserasplysasn
580585590
alaalaaspphesertyrtrpilegluglythrproleuvalalaval
595600605
hisglnargsertyrmetleuargleuproglygluglnvalglulys
610615620
glnvalmetglulysargaspgluargpheglnleuhisglnargval
625630635640
lyspheglnileargvalleualaglnilemetargmetalaasnlys
645650655
glntyrglyaspargtrpaspgluleuaspserleulysglnalaval
660665670
gluglnlyslysserproleuaspglnthraspargthrphetrpglu
675680685
glyilevalcysaspleuthrlysvalleuproargasnglualaasp
690695700
trpgluglnalavalvalglnilehisarglysalagluglutyrval
705710715720
glylysalavalglnalatrparglysargphealaalaaspgluarg
725730735
lysglyilealaglyleusermettrpasnileglugluleuglugly
740745750
leuarglysleuleuilesertrpserargargthrargasnprogln
755760765
gluvalasnargphegluargglyhisthrserhisglnargleuleu
770775780
thrhisileglnasnvallysgluaspargleulysglnleuserhis
785790795800
alailevalmetthralaleuglytyrvaltyraspgluarglysgln
805810815
glutrpcysalaglutyrproalacysglnvalileleuphegluasn
820825830
leuserglntyrargserasnleuaspargserthrlysgluasnser
835840845
thrleumetlystrpalahisargserileprolystyrvalhismet
850855860
glnalagluprotyrglyileglnileglyaspvalargalaglutyr
865870875880
serserargphetyralalysthrglythrproglyileargcyslys
885890895
lysvalargglyglnaspleuglnglyargargphegluasnleugln
900905910
lysargleuvalasngluglnpheleuthrglugluglnvallysgln
915920925
leuargproglyaspilevalproaspaspserglygluleuphemet
930935940
thrleuthraspglyserglyserlysgluvalvalpheleuglnala
945950955960
aspileasnalaalahisasnleuglnlysargphetrpglnargtyr
965970975
asngluleuphelysvalsercysargvalilevalargaspgluglu
980985990
glutyrleuvalprolysthrlysservalglnalalysleuglylys
99510001005
glyleuphevallyslysseraspthralatrplysaspvaltyr
101010151020
valtrpaspserglnalalysleulysglylysthrthrphethr
102510301035
gluglusergluserprogluglnleugluasppheglngluile
104010451050
ilegluglualagluglualalysglythrtyrargthrleuphe
105510601065
argaspproserglyvalphepheprogluservaltrptyrpro
107010751080
glnlysaspphetrpglygluvallysarglysleutyrglylys
108510901095
leuarggluargpheleuthrlysalaarg
11001105
<210>6
<211>1112
<212>prt
<213>bacillus
<400>6
metalaileargserilelysleulysmetlysthrasnserglythr
151015
aspseriletyrleuarglysalaleutrpargthrhisglnleuile
202530
asngluglyilealatyrtyrmetasnleuleuthrleutyrarggln
354045
glualaileglyasplysthrlysglualatyrglnalagluleuile
505560
asnileileargasnglnglnargasnasnglyserserglugluhis
65707580
glyseraspglngluileleualaleuleuargglnleutyrgluleu
859095
ileileproserserileglygluserglyaspalaasnglnleugly
100105110
asnlyspheleutyrproleuvalaspproasnserglnserglylys
115120125
glythrserasnalaglyarglysproargtrplysargleulysglu
130135140
gluglyasnproasptrpgluleuglulyslyslysaspglugluarg
145150155160
lysalalysaspprothrvallysilepheaspasnleuasnlystyr
165170175
glyleuleuproleupheproleuphethrasnileglnlysaspile
180185190
glutrpleuproleuglylysargglnservalarglystrpasplys
195200205
aspmetpheileglnalailegluargleuleusertrpglusertrp
210215220
asnargargvalalaaspglutyrlysglnleulysglulysthrglu
225230235240
sertyrtyrlysgluhisleuthrglyglygluglutrpileglulys
245250255
ilearglyspheglulysgluargasnmetgluleuglulysasnala
260265270
phealaproasnaspglytyrpheilethrserargglnilearggly
275280285
trpaspargvaltyrglulystrpserlysleuprogluseralaser
290295300
proglugluleutrplysvalvalalagluglnglnasnlysmetser
305310315320
gluglypheglyaspprolysvalpheserpheleualaasnargglu
325330335
asnargaspiletrpargglyhissergluargiletyrhisileala
340345350
alatyrasnglyleuglnlyslysleuserargthrlysgluglnala
355360365
thrphethrleuproaspalailegluhisproleutrpileargtyr
370375380
gluserproglyglythrasnleuasnleuphelysleugluglulys
385390395400
glnlyslysasntyrtyrvalthrleuserlysileiletrpproser
405410415
gluglulystrpileglulysgluasnilegluileproleualapro
420425430
serileglnpheasnargglnilelysleulysglnhisvallysgly
435440445
lysglngluileserpheserasptyrserserargileserleuasp
450455460
glyvalleuglyglyserargileglnpheasnarglystyrilelys
465470475480
asnhislysgluleuleuglygluglyaspileglyprovalphephe
485490495
asnleuvalvalaspvalalaproleuglngluthrargasnglyarg
500505510
leuglnserproileglylysalaleulysvalileserseraspphe
515520525
serlysvalileasptyrlysprolysgluleumetasptrpmetasn
530535540
thrglyseralaserasnserpheglyvalalaserleuleuglugly
545550555560
metargvalmetserileaspmetglyglnargthrseralaserval
565570575
serilephegluvalvallysgluleuprolysaspglngluglnlys
580585590
leuphetyrserileasnaspthrgluleuphealailehislysarg
595600605
serpheleuleuasnleuproglygluvalvalthrlysasnasnlys
610615620
glnglnargglngluargarglyslysargglnphevalargsergln
625630635640
ileargmetleualaasnvalleuargleugluthrlyslysthrpro
645650655
aspgluarglyslysalailehislysleumetgluilevalglnser
660665670
tyraspsertrpthralaserglnlysgluvaltrpglulysgluleu
675680685
asnleuleuthrasnmetalaalapheasnaspgluiletrplysglu
690695700
serleuvalgluleuhishisargilegluprotyrvalglyglnile
705710715720
valserlystrparglysglyleusergluglyarglysasnleuala
725730735
glyilesermettrpasnileaspgluleugluaspthrargargleu
740745750
leuilesertrpserlysargserargthrproglyglualaasnarg
755760765
ilegluthraspglupropheglyserserleuleuglnhisilegln
770775780
asnvallysaspaspargleulysglnmetalaasnleuileilemet
785790795800
thralaleuglyphelystyrasplysgluglulysaspargtyrlys
805810815
argtrplysgluthrtyrproalacysglnileileleuphegluasn
820825830
leuasnargtyrleupheasnleuaspargserargarggluasnser
835840845
argleumetlystrpalahisargserileproargthrvalsermet
850855860
glnglyglumetpheglyleuglnvalglyaspvalargserglutyr
865870875880
serserargphehisalalysthrglyalaproglyileargcyshis
885890895
alaleuthrglugluaspleulysalaglyserasnthrleulysarg
900905910
leuilegluaspglypheileasnglusergluleualatyrleulys
915920925
lysglyaspileileproserglnglyglygluleuphevalthrleu
930935940
serlysargtyrlyslysaspseraspasnasngluleuthrvalile
945950955960
hisalaaspileasnalaalaglnasnleuglnlysargphetrpgln
965970975
glnasnsergluvaltyrargvalprocysglnleualaargmetgly
980985990
gluasplysleutyrileprolysserglnthrgluthrilelyslys
99510001005
tyrpheglylysglyserphevallysasnasnthrgluglnglu
101010151020
valtyrlystrpglulysserglulysmetlysilelysthrasp
102510301035
thrthrpheaspleuglnaspleuaspglyphegluaspileser
104010451050
lysthrilegluleualaglngluglnglnlyslystyrleuthr
105510601065
metpheargaspproserglytyrphepheasnasngluthrtrp
107010751080
argproglnlysglutyrtrpserilevalasnasnileilelys
108510901095
sercysleulyslyslysileleuserasnlysvalgluleu
110011051110
<210>7
<211>1149
<212>prt
<213>desulfovibrioinopinatus
<400>7
metprothrargthrileasnleulysleuvalleuglylysasnpro
151015
gluasnalathrleuargargalaleupheserthrhisargleuval
202530
asnglnalathrlysargilegluglupheleuleuleucysarggly
354045
glualatyrargthrvalaspasngluglylysglualagluilepro
505560
arghisalavalglngluglualaleualaphealalysalaalagln
65707580
arghisasnglycysileserthrtyrgluaspglngluileleuasp
859095
valleuargglnleutyrgluargleuvalproservalasngluasn
100105110
asnglualaglyaspalaglnalaalaasnalatrpvalserproleu
115120125
metseralaglusergluglyglyleuservaltyrasplysvalleu
130135140
aspproproprovaltrpmetlysleulysgluglulysalaprogly
145150155160
trpglualaalaserglniletrpileglnseraspgluglyglnser
165170175
leuleuasnlysproglyserproproargtrpilearglysleuarg
180185190
serglyglnprotrpglnaspaspphevalseraspglnlyslyslys
195200205
glnaspgluleuthrlysglyasnalaproleuilelysglnleulys
210215220
glumetglyleuleuproleuvalasnprophephearghisleuleu
225230235240
aspprogluglylysglyvalserprotrpaspargleualavalarg
245250255
alaalavalalahispheilesertrpglusertrpasnhisargthr
260265270
argalaglutyrasnserleulysleuargargaspgluphegluala
275280285
alaseraspgluphelysaspaspphethrleuleuargglntyrglu
290295300
alalysarghisserthrleulysserilealaleualaaspaspser
305310315320
asnprotyrargileglyvalargserleuargalatrpasnargval
325330335
arggluglutrpileasplysglyalathrglugluglnargvalthr
340345350
ileleuserlysleuglnthrglnleuargglylyspheglyasppro
355360365
aspleupheasntrpleualaglnasparghisvalhisleutrpser
370375380
proargaspservalthrproleuvalargileasnalavalasplys
385390395400
valleuargargarglysprotyralaleumetthrphealahispro
405410415
argphehisproargtrpileleutyrglualaproglyglyserasn
420425430
leuargglntyralaleuaspcysthrgluasnalaleuhisilethr
435440445
leuproleuleuvalaspaspalahisglythrtrpileglulyslys
450455460
ileargvalproleualaproserglyglnileglnaspleuthrleu
465470475480
glulysleuglulyslyslysasnargleutyrtyrargserglyphe
485490495
glnglnphealaglyleualaglyglyalagluvalleuphehisarg
500505510
protyrmetgluhisaspgluargserglugluserleuleugluarg
515520525
proglyalavaltrpphelysleuthrleuaspvalalathrglnala
530535540
proproasntrpleuaspglylysglyargvalargthrproproglu
545550555560
valhishisphelysthralaleuserasnlysserlyshisthrarg
565570575
thrleuglnproglyleuargvalleuservalaspleuglymetarg
580585590
thrphealasercysservalphegluleuilegluglylysproglu
595600605
thrglyargalapheprovalalaaspgluargsermetaspserpro
610615620
asnlysleutrpalalyshisgluargserphelysleuthrleupro
625630635640
glygluthrproserarglysglugluglugluargserilealaarg
645650655
alagluiletyralaleulysargaspileglnargleulysserleu
660665670
leuargleuglyglugluaspasnaspasnargargaspalaleuleu
675680685
gluglnphephelysglytrpglyglugluaspvalvalproglygln
690695700
alapheproargserleupheglnglyleuglyalaalaprophearg
705710715720
serthrprogluleutrpargglnhiscysglnthrtyrtyrasplys
725730735
alaglualacysleualalyshisileserasptrparglysargthr
740745750
argproargprothrserargglumettrptyrlysthrargsertyr
755760765
hisglyglylysseriletrpmetleuglutyrleuaspalavalarg
770775780
lysleuleuleusertrpserleuargglyargthrtyrglyalaile
785790795800
asnargglnaspthralaargpheglyserleualaserargleuleu
805810815
hishisileasnserleulysgluaspargilelysthrglyalaasp
820825830
serilevalglnalaalaargglytyrileproleuprohisglylys
835840845
glytrpgluglnargtyrgluprocysglnleuileleuphegluasp
850855860
leualaargtyrargpheargvalaspargproargarggluasnser
865870875880
glnleumetglntrpasnhisargalailevalalagluthrthrmet
885890895
glnalagluleutyrglyglnilevalgluasnthralaalaglyphe
900905910
serserargphehisalaalathrglyalaproglyvalargcysarg
915920925
pheleuleugluargasppheaspasnaspleuprolysprotyrleu
930935940
leuarggluleusertrpmetleuglyasnthrlysvalgluserglu
945950955960
gluglulysleuargleuleuserglulysileargproglyserleu
965970975
valprotrpaspglyglygluglnphealathrleuhisprolysarg
980985990
glnthrleucysvalilehisalaaspmetasnalaalaglnasnleu
99510001005
glnargargphepheglyargcysglyglualapheargleuval
101010151020
cysglnprohisglyaspaspvalleuargleualaserthrpro
102510301035
glyalaargleuleuglyalaleuglnglnleugluasnglygln
104010451050
glyalaphegluleuvalargaspmetglyserthrserglnmet
105510601065
asnargphevalmetlysserleuglylyslyslysilelyspro
107010751080
leuglnaspasnasnglyaspaspgluleugluaspvalleuser
108510901095
valleuproglugluaspaspthrglyargilethrvalphearg
110011051110
aspserserglyilephepheprocysasnvaltrpileproala
111511201125
lysglnphetrpproalavalargalametiletrplysvalmet
113011351140
alaserhisserleugly
1145
<210>8
<211>1090
<212>prt
<213>laceyellasediminis
<400>8
metserileargserphelysleulysilelysthrlysserglyval
151015
asnalaglugluleuargargglyleutrpargthrhisglnleuile
202530
asnaspglyilealatyrtyrmetasntrpleuvalleuleuarggln
354045
gluaspleupheileargasnglugluthrasngluileglulysarg
505560
serlysglugluileglnglygluleuleugluargvalhislysgln
65707580
glnglnargasnglntrpserglygluvalaspaspglnthrleuleu
859095
glnthrleuarghisleutyrglugluilevalproservalilegly
100105110
lysserglyasnalaserleulysalaargphepheleuglyproleu
115120125
valaspproasnasnlysthrthrlysaspvalserlysserglypro
130135140
thrprolystrplyslysmetlysaspalaglyaspproasntrpval
145150155160
glnglutyrglulystyrmetalagluargglnthrleuvalargleu
165170175
gluglumetglyleuileproleuphepromettyrthraspgluval
180185190
glyaspilehistrpleuproglnalaserglytyrthrargthrtrp
195200205
aspargaspmetpheglnglnalailegluargleuleusertrpglu
210215220
sertrpasnargargvalarggluargargalaglnpheglulyslys
225230235240
thrhisaspphealaserargphesergluseraspvalglntrpmet
245250255
asnlysleuargglutyrglualaglnglnglulysserleugluglu
260265270
asnalaphealaproasngluprotyralaleuthrlyslysalaleu
275280285
argglytrpgluargvaltyrhissertrpmetargleuaspserala
290295300
alasergluglualatyrtrpglngluvalalathrcysglnthrala
305310315320
metargglyglupheglyaspproalailetyrglnpheleualagln
325330335
lysgluasnhisaspiletrpargglytyrprogluargvalileasp
340345350
phealagluleuasnhisleuglnarggluleuargargalalysglu
355360365
aspalathrphethrleuproaspservalasphisproleutrpval
370375380
argtyrglualaproglyglythrasnilehisglytyraspleuval
385390395400
glnaspthrlysargasnleuthrleuileleuasplyspheileleu
405410415
proaspgluasnglysertrphisgluvallyslysvalpropheser
420425430
leualalysserlysglnphehisargglnvaltrpleuglngluglu
435440445
glnlysglnlyslysarggluvalvalphetyrasptyrserthrasn
450455460
leuprohisleuglythrleualaglyalalysleuglntrpasparg
465470475480
asnpheleuasnlysargthrglnglnglnileglugluthrglyglu
485490495
ileglylysvalphepheasnileservalaspvalargproalaval
500505510
gluvallysasnglyargleuglnasnglyleuglylysalaleuthr
515520525
valleuthrhisproaspglythrlysilevalthrglytrplysala
530535540
gluglnleuglulystrpvalglygluserglyargvalserserleu
545550555560
glyleuaspserleusergluglyleuargvalmetserileaspleu
565570575
glyglnargthrseralathrvalservalphegluilethrlysglu
580585590
alaproaspasnprotyrlysphephetyrglnleugluglythrglu
595600605
leuphealavalhisglnargserpheleuleualaleuproglyglu
610615620
asnproproglnlysilelysglnmetarggluileargtrplysglu
625630635640
argasnargilelysglnglnvalaspglnleuseralaileleuarg
645650655
leuhislyslysvalasngluaspgluargileglnalaileasplys
660665670
leuleuglnlysvalalasertrpglnleuasnglugluilealathr
675680685
alatrpasnglnalaleuserglnleutyrserlysalalysgluasn
690695700
aspleuglntrpasnglnalailelysasnalahishisglnleuglu
705710715720
provalvalglylysglnileserleutrparglysaspleuserthr
725730735
glyargglnglyilealaglyleuserleutrpserileglugluleu
740745750
glualathrlyslysleuleuthrargtrpserlysargserargglu
755760765
proglyvalvallysargilegluargphegluthrphealalysgln
770775780
ileglnhishisileasnglnvallysgluasnargleulysglnleu
785790795800
alaasnleuilevalmetthralaleuglytyrlystyraspglnglu
805810815
glnlyslystrpilegluvaltyrproalacysglnvalvalleuphe
820825830
gluasnleuargsertyrargphesertyrgluargserargargglu
835840845
asnlyslysleumetglutrpserhisargserileprolysleuval
850855860
glnmetglnglygluleupheglyleuglnvalalaaspvaltyrala
865870875880
alatyrserserargtyrhisglyargthrglyalaproglyilearg
885890895
cyshisalaleuthrglualaaspleuargasngluthrasnileile
900905910
hisgluleuileglualaglypheilelysglugluhisargprotyr
915920925
leuglnglnglyaspleuvalprotrpserglyglygluleupheala
930935940
thrleuglnlysprotyraspasnproargileleuthrleuhisala
945950955960
aspileasnalaalaglnasnileglnlysargphetrphisproser
965970975
mettrppheargvalasncysgluservalmetgluglygluileval
980985990
thrtyrvalprolysasnlysthrvalhislyslysglnglylysthr
99510001005
pheargphevallysvalgluglyseraspvaltyrglutrpala
101010151020
lystrpserlysasnargasnlysasnthrpheserserilethr
102510301035
gluarglysproprosersermetileleupheargaspproser
104010451050
glythrphephelysgluglnglutrpvalgluglnlysthrphe
105510601065
trpglylysvalglnsermetileglnalatyrmetlyslysthr
107010751080
ilevalglnargmetgluglu
10851090
<210>9
<211>1119
<212>prt
<213>spirochaetes
<400>9
metserphethrilesertyrprophelysleuileilelysasnlys
151015
aspglualalysalaleuleuaspthrhisglntyrmetasnglugly
202530
vallystyrtyrleuglulysleuleumetpheargglnglulysile
354045
pheileglygluaspgluthrglylysargiletyrileglugluthr
505560
glutyrlyslysglnileglugluphetyrleuilelyslysthrglu
65707580
leuglyargasnleuthrleuthrleuaspgluphelysthrleumet
859095
arggluleutyrilecysleuvalsersersermetgluasnlyslys
100105110
glypheproasnalaglnglnalaserleuasnilepheserproleu
115120125
pheaspalagluserlysglytyrileleulysglugluasnasnasn
130135140
ileserleuilehislysasptyrglylysileleuleulysargleu
145150155160
argaspasnasnleuileproilephethrlysphethraspilelys
165170175
lysilethralalysleuserprothralaleuaspargmetilephe
180185190
alaglnalaileglulysleuleusertyrglusertrpcyslysleu
195200205
metilelysgluargpheasplysgluvallysilelysgluleuglu
210215220
asnlyscysgluasnlysglngluargasplysilephegluileleu
225230235240
glulystyrgluglugluargglnlysthrphegluglnaspsergly
245250255
phealalyslysglylysphetyrilethrglyargmetleulysgly
260265270
pheaspgluilelysglulystrpleulysglulysaspargserglu
275280285
glnasnleuileasnileleuasnlystyrglnthraspasnserlys
290295300
leuvalglyaspargasnleupheglupheileilelysleugluasn
305310315320
glncysleutrpasnglyaspileasptyrleulysilelysargasp
325330335
ileasnlysasnglniletrpleuaspargproglumetproargphe
340345350
thrmetproaspphelyslyshisproleutrptyrargtyrgluasp
355360365
proserasnserasnpheargasntyrlysilegluvalvallysasp
370375380
gluasntyrilethrileproleuilethrgluargasnasnglutyr
385390395400
pheglugluasntyrthrpheasnleualalysleulyslysleuser
405410415
gluasnilethrpheileprolysserlysasnlysgluphegluphe
420425430
ileaspserasnaspgluglugluasplyslysaspglnlyslysser
435440445
lysglntyrilelystyrcysaspthralalysasnthrsertyrgly
450455460
lysserglyglyileargleutyrpheasnargasngluleugluasn
465470475480
tyrlysaspglylyslysmetaspsertyrthrvalphethrleuser
485490495
ileargasptyrlysserleuphealalysglulysleuglnprogln
500505510
ilepheasnthrvalaspasnlysilethrserleulysileglnlys
515520525
lyspheglyasnglugluglnthrasnpheleusertyrphethrgln
530535540
asnglnilethrlyslysasptrpmetaspglulysthrpheglnasn
545550555560
vallysgluleuasngluglyileargvalleuservalaspleugly
565570575
glnargphephealaalavalsercysphegluilemetsergluile
580585590
aspasnasnlysleuphepheasnleuasnaspglnasnhislysile
595600605
ileargileasnasplysasntyrtyralalyshisiletyrserlys
610615620
thrilelysleuserglygluaspaspaspleutyrlysgluarglys
625630635640
ileasnlysasntyrlysleusertyrglngluarglysasnlysile
645650655
glyilephethrargglnileasnlysleuasnglnleuleulysile
660665670
ileargasnaspgluileasplysglulysphelysgluleuileglu
675680685
thrthrlysargtyrvallysasnthrtyrasnaspglyileileasp
690695700
trpasnasnvalaspasnlysileleusertyrgluasnlysgluasp
705710715720
valileasnleuhislysgluleuasplyslysleugluileaspphe
725730735
lysglupheileargglucysarglysproilepheargserglygly
740745750
leusermetglnargileasppheleuglulysleuasnlysleulys
755760765
arglystrpvalalaargthrglnlysseralagluserilevalleu
770775780
thrprolyspheglytyrlysleulysgluhisileasngluleulys
785790795800
aspasnargvallysglnglyvalasntyrileleumetthralaleu
805810815
glytyrilelysaspasngluilelysasnaspserlyslyslysgln
820825830
lysgluasptrpvallyslysasnargalacysglnileileleumet
835840845
glulysleuthrglutyrthrphealagluaspargproarggluglu
850855860
asnserlysleuargmettrpserhisargglnilepheasnpheleu
865870875880
glnglnlysalaserleutrpglyileleuvalglyaspvalpheala
885890895
protyrthrserlyscysleuseraspasnasnalaproglyilearg
900905910
cyshisglnvalthrlyslysaspleuileaspasnsertrppheleu
915920925
lysilevalvallysaspaspalaphecysaspleuilegluileasn
930935940
lysgluasnvallysasnlysserilelysileasnaspileleupro
945950955960
leuargglyglygluleuphealaserilelysaspglylysleuhis
965970975
ilevalglnalaaspileasnalaserargasnilealalysargphe
980985990
leuserglnileasnpropheargvalvalleulyslysasplysasp
99510001005
gluthrphehisleulysasngluproasntyrleulysasntyr
101010151020
tyrserileleuasnphevalprothrasnglugluleuthrphe
102510301035
phelysvalglugluasnlysaspilelysprothrlysargile
104010451050
lysmetasplyshisglulysgluserthraspgluglyaspasp
105510601065
tyrserlysasnglnilealaleupheargaspaspserglyile
107010751080
phepheasplysserleutrpvalaspglylysilephetrpser
108510901095
valvallysasnlysmetthrlysleuleuarggluargasnasn
110011051110
lyslysasnglyserlys
1115
<210>10
<211>1142
<212>prt
<213>tuberibacilluscalidus
<400>10
metasnilehisleulysgluleuileargmetalathrlysserphe
151015
ileleulysmetlysthrlysasnasnproglnleuargleuserleu
202530
trplysthrhisgluleupheasnpheglyvalalatyrtyrmetasp
354045
leuleuserleupheargglnlysaspleutyrmethisasnaspglu
505560
aspproasphisprovalvalleulyslysglugluileglngluarg
65707580
leutrpmetlysvalarggluthrglnglnlysasnglyphehisgly
859095
gluvalserlysaspgluvalleugluthrleuargalaleutyrglu
100105110
gluleuvalproseralavalglylysserglyglualaasnglnile
115120125
serasnlystyrleutyrproleuthraspproalaserglnsergly
130135140
lysglythralaasnserglyarglysproargtrplyslysleulys
145150155160
glualaglyaspprosertrplysaspalatyrglulystrpglulys
165170175
gluargglngluaspprolysleulysileleualaalaleuglnser
180185190
pheglyleuileproleupheargprophethrgluasnasphislys
195200205
alavalileservallystrpmetprolysserlysasnglnserval
210215220
arglyspheasplysaspmetpheasnglnalailegluargpheleu
225230235240
sertrpglusertrpasnglulysvalalagluasptyrglulysthr
245250255
valseriletyrgluserleuglnlysgluleulysglyileserthr
260265270
lysalaphegluilemetgluargvalglulysalatyrglualahis
275280285
leuarggluilethrpheserasnserthrtyrargileglyasnarg
290295300
alaileargglytrpthrgluilevallyslystrpmetlysleuasp
305310315320
proseralaproglnglyasntyrleuaspvalvallysasptyrgln
325330335
argarghisproarggluserglyaspphelysleuphegluleuleu
340345350
serargprogluasnglnalaalatrpargglutyrproglupheleu
355360365
proleutyrvallystyrarghisalagluglnargmetlysthrala
370375380
lyslysglnalathrphethrleucysaspproilearghisproleu
385390395400
trpvalargtyrglugluargserglythrasnleuasnlystyrarg
405410415
leuilemetasnglulysglulysvalvalglnpheaspargleuile
420425430
cysleuasnalaaspglyhistyrglugluglngluaspvalthrval
435440445
proleualaproserglnglnpheaspaspglnilelyspheserser
450455460
gluaspthrglylysglylyshisasnphesertyrtyrhislysgly
465470475480
ileasntyrgluleulysglythrleuglyglyalaargileglnphe
485490495
asparggluhisleuleuargargglnglyvallysalaglyasnval
500505510
glyargilepheleuasnvalthrleuasnilegluprometglnpro
515520525
pheserargserglyasnleuglnthrservalglylysalaleulys
530535540
valtyrvalaspglytyrprolysvalvalasnphelysprolysglu
545550555560
leuthrgluhisilelysgluserglulysasnthrleuthrleugly
565570575
valgluserleuprothrglyleuargvalmetservalaspleugly
580585590
glnargglnalaalaalaileserilephegluvalvalserglulys
595600605
proaspaspasnlysleuphetyrprovallysaspthraspleuphe
610615620
alavalhisargthrserpheasnilelysleuproglyglulysarg
625630635640
thrgluargargmetleugluglnglnlysargaspglnalailearg
645650655
aspleuserarglysleulyspheleulysasnvalleuasnmetgln
660665670
lysleuglulysthraspgluargglulysargvalasnargtrpile
675680685
lysasparggluarggluglugluasnprovaltyrvalglngluphe
690695700
glumetileserlysvalleutyrserprohisservaltrpvalasp
705710715720
glnleulysserilehisarglysleuglugluglnleuglylysglu
725730735
ileserlystrpargglnserileserglnglyargglnglyvaltyr
740745750
glyileserleulysasnilegluaspileglulysthrargargleu
755760765
leupheargtrpsermetargprogluasnproglygluvallysgln
770775780
leuglnproglygluargphealaileaspglnglnasnhisleuasn
785790795800
hisleulysaspaspargilelyslysleualaasnglnilevalmet
805810815
thralaleuglytyrargtyraspglylysarglyslystrpileala
820825830
lyshisproalacysglnleuvalleuphegluaspleuserargtyr
835840845
alaphetyraspgluargserargleugluasnargasnleumetarg
850855860
trpserargarggluileprolysglnvalalaglnileglyglyleu
865870875880
tyrglyleuleuvalglygluvalglyalaglntyrserserargphe
885890895
hisalalysserglyalaproglyileargcysargvalvallysglu
900905910
hisgluleutyrilethrgluglyglyglnlysvalargasnglnlys
915920925
pheleuaspserleuvalgluasnasnileilegluproaspaspala
930935940
argargleugluproglyaspleuileargaspglnglyglyasplys
945950955960
phealathrleuaspgluargglygluleuvalilethrhisalaasp
965970975
ileasnalaalaglnasnleuglnlysargphetrpthrargthrhis
980985990
glyleutyrargileargcysgluserarggluilelysaspalaval
99510001005
valleuvalproserasplysaspglnlysglulysmetgluasn
101010151020
leupheglyileglytyrleuglnprophelysglngluasnasp
102510301035
valtyrlystrpvallysglyglulysilelysglylyslysthr
104010451050
serserglnseraspasplysgluleuvalsergluileleugln
105510601065
glualaservalmetalaaspgluleulysglyasnarglysthr
107010751080
leupheargaspproserglytyrvalpheprolysaspargtrp
108510901095
tyrthrglyglyargtyrpheglythrleugluhisleuleulys
110011051110
arglysleualagluargargleupheaspglyglyserserarg
111511201125
argglyleupheasnglythraspserasnthrasnvalglu
113011351140
<210>11
<211>137
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>11
gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcc60
cgttgaacttctcaaaaagaacgctcgctcagtgttctgacgtcggatcactgagcgagc120
gatctgagaagtggcac137
<210>12
<211>141
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>12
aactgtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaa60
agcccgttgaacttctcaaaaagaacgctcgctcagtgttctgacgtcggatcactgagc120
gagcgatctgagaagtggcac141
<210>13
<211>139
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>13
ctgtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaag60
cccgttgaacttctcaaaaagaacgctcgctcagtgttctgacgtcggatcactgagcga120
gcgatctgagaagtggcac139
<210>14
<211>127
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>14
gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcc60
cgttgaacttctcaaaaagaacgctcgctcagtgttatcactgagcgagcgatctgagaa120
gtggcac127
<210>15
<211>99
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>15
gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcc60
cgttgaacttctcaaaaagaacgatctgagaagtggcac99
<210>16
<211>93
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>16
gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcc60
cgttgaacttctcaaaaagctgagaagtggcac93
<210>17
<211>91
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>17
gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcc60
cgttgaacttctcaaagctgagaagtggcac91
<210>18
<211>91
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>18
gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcc60
cgttgaacttctcaaaactgagaagtggcac91
<210>19
<211>89
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>19
gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcc60
cgttgaacttctcaagcgagaagtggcac89
<210>20
<211>87
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>20
gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcc60
cgttgaacttctaagcagaagtggcac87
<210>21
<211>85
<212>dna
<213>artificialsequence
<220>
<223>optimizedsgrnascaffold
<400>21
gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcc60
cgttgaacttcaagcgaagtggcac85
<210>22
<211>137
<212>dna
<213>artificialsequence
<220>
<223>aasgrna_scaffold
<400>22
gtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagcc60
cgttgaacttctcaaaaagaacgctcgctcagtgttctgacgtcggatcactgagcgagc120
gatctgagaagtggcac137
<210>23
<211>137
<212>dna
<213>artificialsequence
<220>
<223>aksgrna1_scaffold
<400>23
tcgtctataggacggcgaggacaacgggaagtgccaatgtgctctttccaagagcaaaca60
ccccgttggcttcaagatgaccgctcgctcagcgatctgacaacggatcgctgagcgagc120
ggtctgagaagtggcac137
<210>24
<211>145
<212>dna
<213>artificialsequence
<220>
<223>amsgrna1_scaffold
<400>24
ggaattgccgatctataggacggcagattcaacgggatgtgccaatgcactctttccagg60
agtgaacaccccgttggcttcaacatgatcgcccgctcaacggtccgatgtcggatcgtt120
gagcgggcgatctgagaagtggcac145
<210>25
<211>141
<212>dna
<213>artificialsequence
<220>
<223>bhsgrna_scaffold
<400>25
gaggttctgtcttttggtcaggacaaccgtctagctataagtgctgcagggtgtgagaaa60
ctcctattgctggacgatgtctcttttatttcttttttcttggatgtccaagaaaaaaga120
aatgatacgaggcattagcac141
<210>26
<211>132
<212>dna
<213>artificialsequence
<220>
<223>bssgrna_scaffold
<400>26
ccataagtcgacttacatatccgtgcgtgtgcattatgggcccatccacaggtctattcc60
cacggataatcacgactttccactaagctttcgaatgttcgaaagcttagtggaaagctt120
cgtggttagcac132
<210>27
<211>130
<212>dna
<213>artificialsequence
<220>
<223>bs3sgrna_scaffold
<400>27
ggtgacctatagggtcaatgaatctgtgcgtgtgccataagtaattaaaaattacccacc60
acaggattatcttatttctgctaagtgtttagttgcctgaatacttagcagaaataatga120
tgattggcac130
<210>28
<211>118
<212>dna
<213>artificialsequence
<220>
<223>lssgrna_scaffold
<400>28
ggcaaagaatactgtgcgtgtgctaaggatggaaaaaatccattcaaccacaggattaca60
ttatttatctaatcacttaaatctttaagtgattagatgaattaaatgtgattagcac118
<210>29
<211>126
<212>dna
<213>artificialsequence
<220>
<223>sbsgrna_scaffold
<400>29
gtcttagggtatatcccaaatttgtcttagtatgtgcattgcttacagcgacaactaagg60
tttgtttatcttttttttacattgtaagatgttttacattataaaaagaagataatctta120
ttgcac126
<210>30
<211>86
<212>dna
<213>artificialsequence
<220>
<223>artsgrna1scaffold
<400>30
ggtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagc60
ccgttgaacttcaagcgaagtggcac86
<210>31
<211>84
<212>dna
<213>artificialsequence
<220>
<223>artsgrna2scaffold
<400>31
ggtctaaaggacagaagacaacgggaagtgccaatgtgctctttccaagagcaaacaccc60
cgttgacttcaagcgaagtggcac84
<210>32
<211>79
<212>dna
<213>artificialsequence
<220>
<223>artsgrna3scaffold
<400>32
ggtctaaaggacagaaaatctgtgcgtgtgccataagtaattaaaaattacccaccacag60
acttcaagcgaagtggcac79
<210>33
<211>91
<212>dna
<213>artificialsequence
<220>
<223>artsgrna4scaffold
<400>33
ggtcgtctataggacggcgagtttttcaacgggtgtgccaatggccactttccaggtggc60
aaagcccgttgaacttcaagcgaagtggcac91
<210>34
<211>89
<212>dna
<213>artificialsequence
<220>
<223>artsgrna5scaffold
<400>34
ggtcgtctataggacggcgaggacaacgggaagtgccaatgtgctctttccaagagcaaa60
caccccgttgacttcaagcgaagtggcac89
<210>35
<211>84
<212>dna
<213>artificialsequence
<220>
<223>artsgrna6scaffold
<400>35
ggtcgtctataggacggcgagaatctgtgcgtgtgccataagtaattaaaaattacccac60
cacagacttcaagcgaagtggcac84
<210>36
<211>90
<212>dna
<213>artificialsequence
<220>
<223>artsgrna7scaffold
<400>36
ggtgacctatagggtcaatgtttttcaacgggtgtgccaatggccactttccaggtggca60
aagcccgttgaacttcaagcgaagtggcac90
<210>37
<211>88
<212>dna
<213>artificialsequence
<220>
<223>artsgrna8scaffold
<400>37
ggtgacctatagggtcaatggacaacgggaagtgccaatgtgctctttccaagagcaaac60
accccgttgacttcaagcgaagtggcac88
<210>38
<211>83
<212>dna
<213>artificialsequence
<220>
<223>artsgrna9scaffold
<400>38
ggtgacctatagggtcaatgaatctgtgcgtgtgccataagtaattaaaaattacccacc60
acagacttcaagcgaagtggcac83
<210>39
<211>85
<212>dna
<213>artificialsequence
<220>
<223>artsgrna10scaffold
<400>39
ggtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagc60
ccgttgagcttcaaagaagtggcac85
<210>40
<211>83
<212>dna
<213>artificialsequence
<220>
<223>artsgrna11scaffold
<400>40
ggtctaaaggacagaagacaacgggaagtgccaatgtgctctttccaagagcaaacaccc60
cgttggcttcaaagaagtggcac83
<210>41
<211>78
<212>dna
<213>artificialsequence
<220>
<223>artsgrna12scaffold
<400>41
ggtctaaaggacagaaaatctgtgcgtgtgccataagtaattaaaaattacccaccacag60
gcttcaaagaagtggcac78
<210>42
<211>90
<212>dna
<213>artificialsequence
<220>
<223>artsgrna13scaffold
<400>42
ggtcgtctataggacggcgagtttttcaacgggtgtgccaatggccactttccaggtggc60
aaagcccgttgagcttcaaagaagtggcac90
<210>43
<211>88
<212>dna
<213>artificialsequence
<220>
<223>artsgrna14scaffold
<400>43
ggtcgtctataggacggcgaggacaacgggaagtgccaatgtgctctttccaagagcaaa60
caccccgttggcttcaaagaagtggcac88
<210>44
<211>83
<212>dna
<213>artificialsequence
<220>
<223>artsgrna15scaffold
<400>44
ggtcgtctataggacggcgagaatctgtgcgtgtgccataagtaattaaaaattacccac60
cacaggcttcaaagaagtggcac83
<210>45
<211>89
<212>dna
<213>artificialsequence
<220>
<223>artsgrna16scaffold
<400>45
ggtgacctatagggtcaatgtttttcaacgggtgtgccaatggccactttccaggtggca60
aagcccgttgagcttcaaagaagtggcac89
<210>46
<211>87
<212>dna
<213>artificialsequence
<220>
<223>artsgrna17scaffold
<400>46
ggtgacctatagggtcaatggacaacgggaagtgccaatgtgctctttccaagagcaaac60
accccgttggcttcaaagaagtggcac87
<210>47
<211>82
<212>dna
<213>artificialsequence
<220>
<223>artsgrna18scaffold
<400>47
ggtgacctatagggtcaatgaatctgtgcgtgtgccataagtaattaaaaattacccacc60
acaggcttcaaagaagtggcac82
<210>48
<211>89
<212>dna
<213>artificialsequence
<220>
<223>artsgrna19scaffold
<400>48
ggtctaaaggacagaatttttcaacgggtgtgccaatggccactttccaggtggcaaagc60
ccgttgagattatctatgatgattggcac89
<210>49
<211>87
<212>dna
<213>artificialsequence
<220>
<223>artsgrna20scaffold
<400>49
ggtctaaaggacagaagacaacgggaagtgccaatgtgctctttccaagagcaaacaccc60
cgttggattatctatgatgattggcac87
<210>50
<211>82
<212>dna
<213>artificialsequence
<220>
<223>artsgrna21scaffold
<400>50
ggtctaaaggacagaaaatctgtgcgtgtgccataagtaattaaaaattacccaccacag60
gattatctatgatgattggcac82
<210>51
<211>94
<212>dna
<213>artificialsequence
<220>
<223>artsgrna22scaffold
<400>51
ggtcgtctataggacggcgagtttttcaacgggtgtgccaatggccactttccaggtggc60
aaagcccgttgagattatctatgatgattggcac94
<210>52
<211>92
<212>dna
<213>artificialsequence
<220>
<223>artsgrna23scaffold
<400>52
ggtcgtctataggacggcgaggacaacgggaagtgccaatgtgctctttccaagagcaaa60
caccccgttggattatctatgatgattggcac92
<210>53
<211>87
<212>dna
<213>artificialsequence
<220>
<223>artsgrna24scaffold
<400>53
ggtcgtctataggacggcgagaatctgtgcgtgtgccataagtaattaaaaattacccac60
cacaggattatctatgatgattggcac87
<210>54
<211>93
<212>dna
<213>artificialsequence
<220>
<223>artsgrna25scaffold
<400>54
ggtgacctatagggtcaatgtttttcaacgggtgtgccaatggccactttccaggtggca60
aagcccgttgagattatctatgatgattggcac93
<210>55
<211>91
<212>dna
<213>artificialsequence
<220>
<223>artsgrna26scaffold
<400>55
ggtgacctatagggtcaatggacaacgggaagtgccaatgtgctctttccaagagcaaac60
accccgttggattatctatgatgattggcac91
<210>56
<211>86
<212>dna
<213>artificialsequence
<220>
<223>artsgrna27scaffold
<400>56
ggtgacctatagggtcaatgaatctgtgcgtgtgccataagtaattaaaaattacccacc60
acaggattatctatgatgattggcac86
<210>57
<211>82
<212>dna
<213>artificialsequence
<220>
<223>artsgrna28scaffold
<400>57
ggtctaaaggacagaacaacgggatgtgccaatgcactctttccaggagtgaacaccccg60
ttgacttcaagcgaagtggcac82
<210>58
<211>87
<212>dna
<213>artificialsequence
<220>
<223>artsgrna29scaffold
<400>58
ggtcgtctataggacggcgagcaacgggatgtgccaatgcactctttccaggagtgaaca60
ccccgttgacttcaagcgaagtggcac87
<210>59
<211>99
<212>dna
<213>artificialsequence
<220>
<223>artsgrna30scaffold
<400>59
ggaattgccgatctataggacggcagatttttttcaacgggtgtgccaatggccactttc60
caggtggcaaagcccgttgaacttcaagcgaagtggcac99
<210>60
<211>97
<212>dna
<213>artificialsequence
<220>
<223>artsgrna31scaffold
<400>60
ggaattgccgatctataggacggcagattgacaacgggaagtgccaatgtgctctttcca60
agagcaaacaccccgttgacttcaagcgaagtggcac97
<210>61
<211>95
<212>dna
<213>artificialsequence
<220>
<223>artsgrna32scaffold
<400>61
ggaattgccgatctataggacggcagattcaacgggatgtgccaatgcactctttccagg60
agtgaacaccccgttgacttcaagcgaagtggcac95
<210>62
<211>81
<212>dna
<213>artificialsequence
<220>
<223>artsgrna33scaffold
<400>62
ggtctaaaggacagaacaacgggatgtgccaatgcactctttccaggagtgaacaccccg60
ttggcttcaaagaagtggcac81
<210>63
<211>86
<212>dna
<213>artificialsequence
<220>
<223>artsgrna34scaffold
<400>63
ggtcgtctataggacggcgagcaacgggatgtgccaatgcactctttccaggagtgaaca60
ccccgttggcttcaaagaagtggcac86
<210>64
<211>98
<212>dna
<213>artificialsequence
<220>
<223>artsgrna35scaffold
<400>64
ggaattgccgatctataggacggcagatttttttcaacgggtgtgccaatggccactttc60
caggtggcaaagcccgttgagcttcaaagaagtggcac98
<210>65
<211>96
<212>dna
<213>artificialsequence
<220>
<223>artsgrna36scaffold
<400>65
ggaattgccgatctataggacggcagattgacaacgggaagtgccaatgtgctctttcca60
agagcaaacaccccgttggcttcaaagaagtggcac96
<210>66
<211>94
<212>dna
<213>artificialsequence
<220>
<223>artsgrna37scaffold
<400>66
ggaattgccgatctataggacggcagattcaacgggatgtgccaatgcactctttccagg60
agtgaacaccccgttggcttcaaagaagtggcac94
-
 0访客 来自[重庆市电信] 2020年04月25日 14:33看不懂
0访客 来自[重庆市电信] 2020年04月25日 14:33看不懂