用于治疗癌症的包含向导RNA和核酸内切酶作活性成分的药物组合物的制作方法
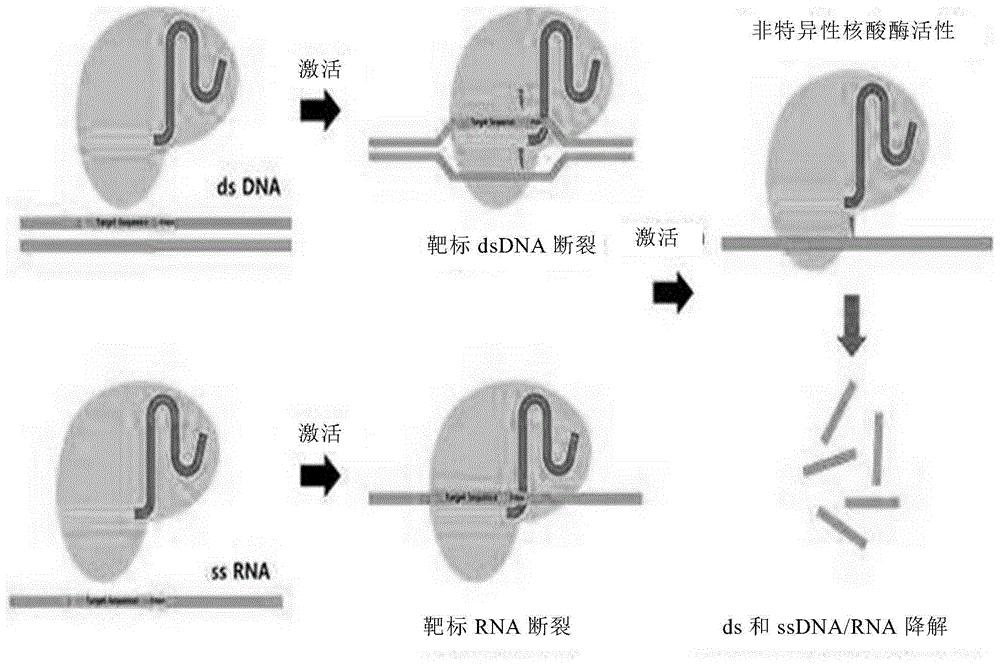
本公开的实施方式涉及包含向导rna和核酸内切酶的抗癌组合物。
背景技术:
:尽管癌症治疗技术有了重大突破,但是癌症仍然是对人类威胁最大的疾病。癌症可由多种致癌物触发,是一种可由染色体结构或dna碱基序列变化引起的疾病。癌症最显著的特征之一是持续的细胞增殖。当前,最广泛使用的抗癌疗法是有效杀伤细胞的放射疗法,或者是使用靶向特定癌细胞的化合物或抗体的疗法。但是,包括化学疗法/放射疗法在内的一线疗法可能还会杀伤体内正常的增殖细胞,例如头发和免疫细胞,从而给患者带来严重的副作用和痛苦。因此,需要开发能够选择性地仅杀死体内癌细胞的抗癌剂。为满足这种需求,针对靶向抗癌剂的研究正在积极进行中。靶向抗癌剂是通过控制参与癌症发展中特定的蛋白或通路来治疗癌症的癌症药物。通过将癌细胞的总蛋白水平与正常细胞的总蛋白水平进行比较,可以确定癌细胞特异的靶标。换句话说,特异存在于癌细胞中或癌细胞中富含的蛋白可能是潜在的靶标。靶蛋白的一个例子是人表皮生长因子受体2蛋白(her-2)。为了治疗过度表达her-2的乳腺癌和胃癌,几种针对her-2的靶向治疗剂已被开发,包括曲妥珠单抗(trastuzumab)另一种方法是靶向导致癌症恶化的突变蛋白。例如,分别以修饰形式存在于许多乳腺癌和黑色素瘤中的细胞增殖信号蛋白brca1和braf。此类突变的许多靶向治疗剂已被开发,并且由此开发的靶向治疗剂已被批准用于治疗无法进行手术或具有转移性癌症的患者。最近的靶向治疗剂和免疫治疗剂是基于在癌细胞中特异性表达的生物标记蛋白研发的。然而,癌细胞中特异性存在的生物标志物蛋白与抗癌剂之间的相互作用强度不是特定的,有时可能会导致副作用。此外,尽管靶向治疗剂和免疫治疗剂的毒性比常规化疗剂低,但仍有许多副作用被报导。因此,在抗癌治疗领域,仍需开发特异性靶向体内癌细胞的抗癌剂。特别地,基于dna序列差异,即分化癌细胞中的最显著特征开发抗癌剂,是人类长期以来的愿景。发明公开内容技术问题癌细胞中存在的染色体结构异常或癌细胞中不同于正常细胞的碱基序列可能是区分癌细胞与正常细胞的重要标准。因此,染色体或碱基序列中的此类差异可能是癌症治疗的重要靶标。因此,实施方式中的一个目的是提供用于治疗癌症的药物组合物,其通过靶向特异性存在于癌细胞中的序列而表现出抗癌作用。技术问题的解决方案为了实现上述目的,一实施方式提供了用于杀伤肿瘤细胞的组合物,该组合物包含与特异性存在于癌细胞中的核酸互补结合的多核苷酸和核酸酶作为活性成分。另外,一实施方式提供了用于治疗癌症的药物组合物,其包含上述组合物。此外,一实施方式提供了用于杀伤肿瘤细胞的组合物,该组合物包含作为活性成分的载体,所述载体包含与特异性存在于癌细胞中的核酸互补结合的多核苷酸以及编码核酸内切酶和核酸外切酶的多核苷酸。进一步,一实施方式提供了治疗癌症的方法,该方法包括将本发明的组合物施用于患有癌症的受试者。发明的有益效果根据该实施方式的药物组合物是基于dna和rna之间单独高特异性的药物,可以针对每个患者和每种癌症进行定制,因为其可以特异性地靶向和杀伤癌细胞。特别地,通过选择性地靶向仅存在于癌细胞中的具有单核苷酸多态性(snp)和/或拷贝数变异(cnv)的基因,可以有效地仅杀伤患者的癌细胞。另外,由于靶基因在正常细胞中不存在,因此仅癌细胞可以被有效地去除。因此,在安全性方面,该实施方式的制剂优于常规抗癌剂。特别地,当使用含有crispr相关蛋白与核酸外切酶,例如recj组合的融合蛋白时,可以提供更优异的抗癌剂。附图说明图1是一示意图,其中crispr相关蛋白的非特异性核酸外切酶功能被crrna引导的靶位点结合激活,从而降解癌细胞的靶核酸。图2是一示意图,显示了根据实施例所述的由crrna、crispr相关蛋白和/或核酸外切酶组成的组合物(在下文中,称为crisprplus系统)在癌细胞中被特异性激活,并起到抗癌剂的作用。图3表明,根据实施例的crispr/cas12a蛋白具有非特异性核酸外切酶活性,这取决于其序列特异性内切酶活性。图4显示了在人癌细胞株hek293(图4a)和hela(图4b)转染crispr/cas12a(cpf1)核酸酶、crrna或两个分子结合的rnp复合物,在24、48和72小时后测得的细胞活力。图5显示了特异性结合egfr突变体和cas9的向导rna,在仅脉冲、egfr_wt和具有egfr突变体的hcc827细胞株实验组中诱导的细胞凋亡。图6显示的图比较了图5中检测的仅脉冲、egfr_wt和具有egfr突变体的hcc827细胞株实验组的活细胞数。图7显示了与靶基因ccr5、hprt1、mt2、smim11、gnpda2、slc15a5和kcne2互补的向导rna在肺癌细胞系h1299中诱导的肺癌细胞凋亡。在该实验中,进行脂质转染以导入编码向导rna和cas9的核酸。nt1是指不含有与肺癌细胞中的序列互补匹配的序列的向导rna,用作阴性对照。特别地,证实了与rnamt2、smim11、gnpda2、slc15a5和kcne2互补结合的向导rna可以有效地杀伤肺癌细胞。图8显示了为证实图7所示的凋亡结果而测得的活细胞数。图9显示的图证实肺癌细胞系h1299中与靶基因ccr5、hprt1、mt2、gnpda2、slc15a5和kcne2互补的向导rna诱导的肺癌细胞凋亡。包含脂质体的对照组,其中仅进行脂质体转染处理而不导入dna。图10显示了通过nucbluelivereadyprobes试剂和碘化丙啶readyprobes试剂获得的nt1对照和gnpda2实验组的活细胞的显微图像。nucbluelivereadyprobes试剂是一种蓝色荧光染料,可对活细胞和死细胞进行染色。碘化丙啶readyprobes试剂是一种红色荧光染料,仅染色死细胞。图11显示了检测到的活细胞数,以证实肺癌细胞系h1563中与靶基因ccr5、hprt1、mt2、irx1和adamts16互补的向导rna诱导的肺癌细胞凋亡。图12显示了发光法检测的活细胞数量随时间的变化,以证实肺癌细胞系a549中与靶基因hprt1、ccr5和mt2互补的向导rna诱导的肺癌细胞凋亡。图13显示了测定的活细胞数,以证实在肺癌细胞系a549中与靶基因hprt1、ccr5和mt2互补的向导rna诱导的肺癌细胞凋亡。图14显示了图13结果的显微镜观察结果。图15显示了发光法检测的活细胞数量随时间的变化,以证实肺癌细胞系a549中与靶基因mt2、cd68、dach2、herc2p2和shbg互补的向导rna诱导的肺癌细胞凋亡。图16显示了发光法检测的活细胞数量随时间的变化,以证实乳腺癌细胞系skbr3中与靶基因mt2、erbb2和krt16互补的向导rna诱导的乳腺癌细胞凋亡。图17显示了检测的活细胞数量随时间的变化,以证实与乳腺癌细胞系skbr3中的靶基因mt2、erbb2和krt16互补的向导rna诱导的乳腺癌细胞凋亡。图18显示了检测的活细胞数量随时间的变化,以证实与子宫颈癌细胞hela中的靶基因ccr5、mt2和prdm9互补的向导rna诱导的子宫颈癌细胞凋亡。对于ccr5,每个基因的cnv为2,对于mt2,至少为100,对于prdm9,至少为8。图19显示了检测的活细胞数量随时间的变化,以证实与宫颈癌细胞系hela中的靶基因ccr5、mt2、prdm9和hpv_1互补的向导rna诱导的宫颈癌细胞凋亡。对于ccr5,每个基因的cnv是2,对于mt2,至少为100,对于prdm9,至少为8,对于hpv_1为30。图20a显示了检测的活细胞数量随时间的变化,以证实与宫颈癌细胞hela中的靶基因ccr5、mt2、hpv_1和prdm9互补的向导rna诱导的宫颈癌细胞凋亡。图20b示出了检测的活细胞数量随时间的变化,以证实与宫颈癌细胞hela中的靶基因ccr5、mt2和hpv_1互补的向导rna诱导的宫颈癌细胞凋亡。靶向人基因组中不存在区域的nt序列nt1、nt2和nt3作为阴性对照组。图21显示了检测到的活细胞数,以证实与结肠癌细胞系ht-29中与靶基因ccr5、hprt1、mt2、trappc9、linc00536、trps1和cdk8互补的向导rna诱导的结肠癌细胞凋亡。图22显示了发光法检测的活细胞数量随时间的变化,以证实结肠癌细胞系ht-29中与靶基因mt2、cdk8、linc00536、trps1和trappc9互补的向导rna诱导的结肠癌细胞凋亡。图23显示了测定的活细胞数,以证实在结肠癌细胞系h1563中与靶基因smim11、gnpda2、sls15a5和kcne2互补的向导rna诱导的肺癌细胞凋亡。本实验目的是为了验证在肺癌细胞株h1299中有效的与靶基因互补的引导rna是否在其他肺癌细胞株h1563中也有效。图24提供了对图23结果的显微观察结果。图25显示了与靶基因ccr5、kcne2、gnpda2、smim11和sls15a5互补的引导rna诱导的肺癌细胞凋亡图。进行该实验以证实cnv在肺癌细胞系h1299中的作用。由于添加了annv试剂,因此在死细胞中检测到高荧光。图26显示了cas9(具有内切核酸酶功能的crispr相关蛋白)和recj(具有核酸外切酶活性)的融合蛋白、cas9和cas9-recj(seqidno:87)的癌细胞的杀伤作用,通过利用与肺癌系hcc827、中的egfr突变体和ccr5互补的引导rna。结果证实,含有核酸外切酶的融合蛋白对肺癌细胞的杀伤能力明显增强。图27为图26结果的显微观察结果。图28显示了用于确认效果依赖于递送系统的实验结果,其中检测组合了引导rna和内切酶蛋白cas9的rnp(核糖核蛋白)是否具有凋亡作用。具体地,实验证实,与肺癌细胞系hcc827中的egfr突变体互补结合的引导rna与cas9组合的rnp有效地杀伤肺癌细胞。另一方面,用作阴性对照组的cas9蛋白和向导rna没有杀死细胞。图29示出了用于确认效果取决于递送系统的实验结果。在该实验中,检查了rnp是否可以有效诱导细胞凋亡。将sgrna用作对照组,并且将由与mt2和cas9互补的引导rna组成的rnp用作实验组。细胞系是h1563,一种肺癌细胞系。图30示出了确认效果取决于递送系统的实验结果。在该实验中,检查了rnp是否可以有效诱导细胞凋亡。sgrna和cas9蛋白用作对照组,rnps分别由cas9和与ccr5、gnpda2和smim11中任一个互补的引导rna组成,用作实验组。细胞系是h1299,一种肺癌细胞系。图31示出了确认效果取决于递送系统的实验结果。在该实验中,检查了rnp是否可以有效诱导细胞凋亡。sgrna和cas9蛋白用作对照组,rnps分别由cas9和与ccr5或mt2互补的引导rna组成,用作实验组。细胞系是h1299,一种肺癌细胞系。图32提供了对图30结果的显微观察结果。图33显示了使用与靶基因ccr5和egfr_e2互补的引导rna和cas12a诱导的细胞凋亡。该细胞系是hcc827,一种肺癌细胞系。图34为图33结果的显微观察结果。发明实施的最佳方式本公开的实施方式提供了用于杀伤肿瘤细胞的组合物,该组合物包含与特异性存在于癌细胞中的核酸互补结合的多核苷酸和核酸酶作为活性成分。根据一实施方式,多核苷酸可以是crrna或grna。crrna是指crisprrna。另外,grna是指向导rna。crrna和grna可以是单链rna。此外,crrna可以与tracrrna结合以激活crispr相关蛋白,并且crrna可以与tracrrna结合使用。crrna可以具有与特异性存在于靶癌细胞中的基因序列互补的序列。此外,grna可以结合特异性存在于靶癌细胞中的基因序列,从而使crispr相关蛋白表现出活性。crrna或grna可以是由15至40个核苷酸组成的rna。所述多核苷酸可以由18至30或20至25个核苷酸组成。例如,crrna或grna可以由20个核苷酸组成。此外,crrna或grna可能在3'端包含其他序列,以使crispr相关蛋白(如cas9)具有活性。在一个实施方式中,grna可以是rna,其是由seqidno:87至129所示的dna代表。如本文所用,术语“核酸酶”可以意指核酸内切酶。核酸酶可以是crispr相关蛋白。如本文所用,术语“crispr相关蛋白”是指能够识别和切割双链或单链核酸如dna和rna(dsdna/rna和ssdna/rna)的酶。特别地,它们可以识别并切割与crrna或向导rna结合的双链或单链核酸。在本公开的一实施方式中,本发明的核酸酶可以是通过识别crrna与靶位点的结合而激活其功能的核酸内切酶。此外,由于核酸内切酶功能被激活,其可能具有能够非特异性地切割双链和/或单链dna和/或rna的核酸外切酶活性。同样,与crispr相关的蛋白质(如cas12a)一旦被激活,可能会表现出非特异性核酸外切酶活性。它可以非特异性地切割dna和rna。因此,本公开的示例性组合物可以通过非特异性核酸酶特异性杀死癌细胞,所述非特异性核酸酶由crrna或grna与癌细胞中存在的特定靶位点的结合而被激活。如上所述,来自一个实施方式的组合物能够仅特异性杀伤癌细胞,因此可以用作抗癌剂。例如,与crispr相关的蛋白质可以是选自下组的任何一种核酸酶:cas1、cas1b、cas2、cas3、cas4、cas5、cas6、cas7、cas8、cas9、cas10、cas12a、cas12b、cas12c、cas12d、cas12e、cas12g、cas12h、cas12i、cas13a、cas13b、cas13c、cas13d、cas14、csy1、csy2、csy3、cse1、cse2、csc1、csc2、csa5、csn2、csmt2、csm3、csm4、csm5、csm6、cr4、cmr5、cmr6、csb1、csb2、csb3、csx17、csx14、csx10、csx16、csax、csx3、csx1、csx15、csf1、csf2、csf3和csf4。优选地,与crispr相关的蛋白质可以是cas9、cas12a(cpf1)或cas13a(c2c2)的核酸酶。作为crispr相关蛋白的一个例子,cas9蛋白可以具有如seqidno:20所示的氨基酸序列。cas9蛋白可以由具有seqidno:19所示序列的核酸编码。此外,cpf1蛋白可以有seqidno:22所示的氨基酸序列。cpf1蛋白可以由具有seqidno:21所示序列的核酸编码。此外,c2c2蛋白可以有seqidno:24所示的氨基酸序列。c2c2蛋白可以由具有seqidno:23所示序列的核酸编码。如本文所用,术语“癌细胞中特异性存在的核酸”是指仅存在于癌细胞中的核酸,其将癌细胞与正常细胞区分开。即,它可以指与正常细胞中不同的序列,并且序列可能在至少一个核酸上不同。此外,基因的一部分可以被取代或缺失。而且,它可以具有重复特定序列的序列。在这种情况下,重复序列可以是细胞中存在的序列,也可以是外部插入的序列。例如,癌细胞中特定存在的核酸可以通过单核苷酸多态性(snp)、拷贝数变异(cnv)、结构变异(sv)、基因插入或基因缺失来表征。具体地,癌细胞中特异性存在的序列可以是癌细胞中存在的snp。存在于癌细胞中的具有上述序列的靶标dna和具有与靶标dna互补的序列的crrna或向导rna可以彼此特异性结合。因此,癌细胞中特异性存在的核酸可以赋予对杀伤肿瘤细胞的组合物的特异性。特别地,作为癌细胞中特异性存在的核酸,可以通过各种癌症组织的基因组序列分析来识别仅存在于癌细胞中的特定snp,并且可以使用特定snp制备crrna或grna。因此,由于其表现出癌细胞特异性毒性,因此可以开发患者定制的抗癌治疗剂。另外,在癌细胞中特异性存在的序列可以包含在癌细胞中存在的拷贝数变异(cnv)。cnv是指基因组部分重复的变异。重复基因的数目可以根据癌症类型或个体而变化。常规地,cnv是指与人类参考基因组相比,通过缺失、扩增等在重复序列的数目上显示差异的核酸片段,不同于以2个拷贝数存在的普通基因。例如,在正常细胞中cnv为2但在癌细胞中cnv为4或以上的基因可以赋予组合物杀死特定肿瘤细胞。cnv可以至少为4、8、10、12、14、16、18、20、24、30、40、50、60、70、80、90或100。特别地,当拷贝数是7或更大时,可以将其确定为cnv。每种癌细胞系的基因的拷贝数的具体实例显示在下表1中。[表1]cnv是与人类疾病,如癌症、智力障碍、癫痫、精神分裂症、儿童肥胖等相关的最重要的变异类型之一。大多数癌细胞系具有cnv,并且cnv中存在的靶序列和与其互补的序列的向导rna彼此特异性结合。因此,特异性存在于癌细胞中的cnv可以赋予本公开的抗癌剂特异性。特别地,cnv的数目越高,被crispr相关蛋白切割的基因的数目越多,从而导致癌细胞核的显著损伤。特别是,癌细胞中特异性存在的cnv数据可以通过微阵列和荧光原位杂交(fish)等技术轻松获得,并可用于产生crrna或grna。当用靶向癌细胞cnv中特定序列的crrna或grna和crispr相关蛋白治疗癌细胞时,可能会诱导癌细胞特异性凋亡。因此,由于其表现出癌细胞特异性毒性,因此可以开发患者定制的抗癌治疗剂。在本公开的一个实施例中,可以使用以下等式从拷贝数值(v)计算cnv的预期拷贝数(n):n=2×2v。在本公开的一个实施方式中,已经证实,当互补结合癌细胞中特别高的cnv的基因的多核苷酸与crispr相关蛋白或crisprplus蛋白一起使用时,仅癌细胞可以被有效杀伤,其中crispr相关蛋白与核酸外切酶蛋白融合。此外,已经证实,使用从癌细胞中特有的基因突变中选择具有高cnv的基因可以更有效地杀死癌细胞。此外,癌细胞中特异性存在的序列可以是癌细胞中存在的结构变异(sv),并且sv可以是倒位、易位、短核苷酸重复扩增等。倒位是其中一部分基因被倒置的突变,是与诸如血友病和肺癌等疾病相关的突变类型之一。易位是其中一部分染色体脱落并与另一条染色体结合的突变。短核苷酸重复序列扩增是其中相同序列被连续重复和过度扩增的突变。大多数癌细胞系具有诸如基因倒置、易位和短核苷酸重复扩增等sv,sv的序列末端与正常细胞系的序列末端之间的连接序列仅存在于相应的癌细胞系中。sv连接序列存在于癌细胞中,和具有互补序列的向导rna相互特异性结合。因此,癌细胞中特异性存在的sv连接序列可以赋予对本发明抗癌剂的特异性。特别地,与cnv不同,sv连接序列在正常细胞中不存在,相应地,它对癌细胞具有特异性。因此,当用与癌细胞中存在的sv连接序列互补结合的多核苷酸和crispr相关蛋白处理癌细胞时,可以诱导癌细胞特异性凋亡。因此,由于其仅对特定癌细胞表现出特异性毒性,因此可以开发患者定制的抗癌治疗剂。此外,可以通过基因的插入或缺失来产生癌细胞中特异性存在的序列。具有与通过插入或缺失引入的突变序列互补的序列的向导rna,可以有效杀伤癌细胞。因此,癌细胞中特定存在的基因的插入或缺失可以赋予对本发明抗癌剂的特异性。特别地,插入和缺失突变序列对癌细胞是特异性的,因为它们像sv连接序列一样,不存在于正常细胞中,并且当用crispr相关蛋白处理癌细胞中存在的插入或缺失突变序列时,可能会特异性地诱导凋亡。然而,当靶向插入和缺失突变体序列时,可能需要能被indel(插入和缺失)附近的基因识别的pam(原间隔子相邻基序)序列。另外,插入的基因可以是细胞内存在的核酸序列,也可以是外部导入的基因序列。特别地,在由病毒感染等引起的癌细胞的情况下,可以将病毒基因插入细胞中。在本公开的一个实施方式中,病毒核酸序列可用作特异性存在于癌细胞中的核酸。特别地,具有这种插入突变的癌症并不常见,但是插入的病毒序列是癌细胞特有的,因为它在正常细胞中不存在。另外,当病毒序列以多拷贝整合时,cnv很高,这肯定能诱导癌细胞的凋亡。这种癌症的一个例子可以是由乳头瘤病毒引起的宫颈癌。此类靶向外部导入基因的grna的例子可以是任一个seqidno:121至124所示的dna产生的grna。此外,5'-ngg-3'序列,即pam(原间隔子相邻基序)序列,可一并考虑以选择对癌细胞具有特异性的基因和与该基因互补的多核苷酸序列。例如,当5'-ngg-3'序列存在于癌细胞特异性序列附近时,可以将3'方向上的20个核苷酸指定为靶标。在选择目标基因时,可以优先选择具有明确的序列信息,例如,基因的插入、基因的缺失和连接区域的基因以及具有高cnv拷贝数的基因。为了选择靶序列,可以确认在序列的5'和3'末端是否存在g或c,或者整个序列的gc含量(%)是否在40%至60%之内,以及pam3'方向的第3个至第4个碱基部分(即与核酸内切酶,crispr相关的蛋白质的裂解序列)是a还是t。当g或c出现序列的5'-端和3'-端时,及当整个序列的gc含量(%)在40%到60%之内时,sgrna的结合亲和力可能会增加。特别地,当pam的3’方向上的第三至第四碱基部分为a或t,即酿脓链球菌(streptococcuspyogenes)cas9(spcas9)切割序列位点时,可以提高spcas9的切割效率。上述癌症的例子可以是选自以下癌症的任何一种:膀胱癌、骨癌、血液癌、乳腺癌、黑素瘤、甲状腺癌、甲状旁腺癌、骨髓癌、直肠癌、喉癌、喉部癌、肺癌、食道癌、胰腺癌、胃癌、舌癌、皮肤癌、脑瘤、子宫癌、头颈癌、胆囊癌、口腔癌、结肠癌、肛周癌、中枢神经系统肿瘤、肝癌和结直肠癌。特别地,可能是胃癌、结直肠癌、肝癌、肺癌或乳腺癌,在韩国被称为五种主要癌症。上述癌症中特异性存在的核酸可以是选自以下的任何一种基因的突变体:p53、pten、apc、msh2、hbv、hcv和egfr,但不限于此。具体而言,对于胃癌,它可能是p53或pten的突变体,称为肿瘤抑制基因。在结肠癌的情况下,它可能是apc或msh2基因的突变体。另外,肝癌主要是由hbv和hcv病毒的感染引起的,因此可以靶向hbv或hcv的核酸。另外,在肺癌中,egfr基因的突变可作为靶标,并且在乳腺癌的情况下,brca1/2基因的突变可作为主要靶标。如上所述,可以选择与癌症的发展密切相关的突变基因和病毒基因作为癌细胞中特异性存在的核酸序列,并且可以用于生产crrna或grna。此时,可以使用癌细胞中特有的任何dna的snp。癌细胞中特异性存在的核酸序列的实例可以是下表2中描述的序列,但不限于此。[表2]此时,靶向癌细胞中特异存在的核酸序列的crisprrna可能包含一个或多个crrna或grna序列。例如,可以使用可以同时靶向卵巢癌或乳腺癌中存在的brca1的外显子10或11的crrna或grna。另外,可以使用靶向brca1外显子11的两个或更多个crrna或grna。因此,可以根据癌症治疗的目的和癌症的种类适当地选择crrna或grna的组合。即,可以选择和使用不同的grna。本公开内容的肿瘤杀伤组合物可以进一步包含核酸外切酶。如本文所用,术语“核酸外切酶”是从核酸分子的5'或3'末端切割核苷酸的酶。因此,核酸外切酶可以是在5'至3'方向上降解核酸的5'→3'核酸酶。另外,核酸外切酶可以是在3'至5'方向上降解核酸的3'→5'核酸酶。具体地,5'→3'核酸酶的实例可以是源自大肠杆菌(e.coli)的rece或recj。它也可以是源自噬菌体t5的t5。另外,3'→5'核酸酶的实例可以是源自真核细胞或原核细胞的exoi。也可能是源自大肠杆菌(e.coli)的exoiii它也可能是人源衍生的trex1或trex2。另外,具有5'→3’和3'→5'双向切割活性的核酸酶可以是来源于大肠杆菌(e.coli)的exovii或recbcd。此外,例如,可以是来自大肠杆菌(e.coli)的5'→3’λ核酸外切酶。它也可能是源自辐射豇豆(vignaradiata)的绿豆,可以切割单链dna。示例性的核酸外切酶可以是选自以下的任意一种:核糖核酸外切酶t、trex2、trex1、recbcd、脱氧核糖核酸外切酶i、脱氧核糖核酸外切酶iii、绿豆核酸外切酶、rece、recj、t5、λ核酸外切酶、核酸外切酶vii小单元、核酸外切酶、exoi、exoiii、exovii和lexo。具体地,核酸外切酶可以选自以下的任一种:核糖核酸外切酶t(seqidno:4)、trex2(seqidno:5)、trex1(seqidno:6)、recbcd_recb(seqidno:7)、recbcd_recc(seqidno:8)、recbcd_recd(seqidno:9)、脱氧核糖核酸外切酶i(seqidno:10)、脱氧核糖核酸外切酶iii(seqidno:11)、绿豆核酸外切酶(seqidno:12)、recj(seqidno:13)、rece(seqidno:14)、t5(seqidno:15)、λ核酸外切酶(seqidno:16)、核酸外切酶vii小单位(seqidno:17)和核酸外切酶vii大单元(seqidno:18)。另一个实施方式提供了用于杀伤肿瘤细胞的组合物,其包含与癌细胞中特异存在的核酸互补结合的多核苷酸以及由内切核酸酶和核酸外切酶组成的融合蛋白作为活性成分。术语“在癌细胞中特异性存在的核酸”、“与在癌细胞中特异性存在的核酸互补结合的多核苷酸”、“核酸内切酶”和“核酸外切酶”如上所述。例如,通过将外切核酸酶与crrna和crispr相关蛋白结合制备的能够有效杀伤具有所需核酸序列的细胞的crispr/cas系统被称为crisprplus。由内切核酸酶和核酸外切酶组成的融合蛋白的实例可以是cas9-核糖核酸外切酶t、cas9-rex2、cas9-trex1、cas9-recbcd_recb、cas9-recbcd_recc、cas9-recbcd_recd、cas9-脱氧核糖核酸外切酶i、cas9-脱氧核糖核酸外切酶iii、cas9-绿豆酶、cas9-recj、cas9-rece、cas9-t5、cas9-λ、cas9-核酸外切酶vii小单元、cas9-核酸外切酶vii大单元、cpf1-核糖核酸外切酶t、cpf1-rex2、cpf1-trex1、cpf1-recbcd_recb、cpf1-recbcd_recc、cpf1-recbcd_recd、cpf1-脱氧核糖核酸外切酶i、cpf1-脱氧核糖核酸外切酶iii、cpf1-绿豆酶、cpf1-recj、cpf1-rece、cpf1-t5、cpf1-λ、cpf1-核酸外切酶vii小单元vii大单元,优选cas9-recj或cpf1-recj,但不限于此。在本公开的一个实施方式中,使用包含核酸内切酶和核酸外切酶的融合蛋白的crisprplus蛋白导致少数cnv(例如具有cnv2的ccr5基因)中凋亡率增加。与单独使用核酸内切酶时相比,观察到了凋亡作用。一方面,核酸内切酶和核酸外切酶可以通过接头连接。接头可以是白蛋白接头或肽接头。接头可包含1至50个氨基酸,3至40个氨基酸或10至30个氨基酸。另外,肽接头可以是由gly和ser残基组成的肽。此外,肽接头可以是由1至10个氨基酸组成的肽,所述氨基酸选自亮氨酸(leu,l)、异亮氨酸(ile,i)、丙氨酸(ala,a),缬氨酸(val,v)、脯氨酸(pro,p)、赖氨酸(lys,k),精氨酸(arg,r)、天冬酰胺(asn,n)、丝氨酸(ser,s)和谷氨酰胺(gln,q)。另外,接头可以是由3至15个氨基酸组成的多肽,该氨基酸由甘氨酸(gly,g)和丝氨酸(ser,s)残基组成,并且可以由6至11个氨基酸组成。在另一方面,提供了用于治疗癌症的药物组合物,该药物组合物包括上述杀伤肿瘤细胞的组合物。所述肿瘤是选自以下癌症的任何一种:膀胱癌、骨癌、血液癌、乳腺癌、黑素瘤、甲状腺癌、甲状旁腺癌、骨髓癌、直肠癌、喉癌、喉部癌、肺癌、食道癌、胰腺癌、胃癌、舌癌、皮肤癌、脑瘤、子宫癌、头颈癌、胆囊癌、口腔癌、结肠癌、肛周癌、中枢神经系统肿瘤、肝癌和结直肠癌。本公开的药物组合物的制剂可以是肠胃外的。当配制时,通常使用稀释剂或赋形剂,例如填充剂、增量剂、粘合剂、湿润剂、崩解剂或表面活性剂。特别地,肠胃外给药的制剂包括无菌水溶液、非水溶液、悬浮液、乳剂、冻干制剂和栓剂。作为用于非水溶液和悬浮液的溶剂,可以使用丙二醇、聚乙二醇、植物油例如橄榄油,可注射酯例如油酸乙酯等。本公开的药物组合物可以肠胃外施用,并且可以通过选自肿瘤内、静脉内、肌内、皮内、皮下、腹膜内、小动脉内、心室内、病灶内、鞘内、局部,及其组合。本公开的药物组合物的剂量根据患者的体重、年龄、性别、健康状况、饮食、给药时间、给药方法、排泄率和疾病的严重性而变化,并且可以由本领域技术人员适当地选择。为了获得期望的效果,本发明的药物组合物可以以每天0.01μg/kg至100mg/kg,更具体地,1μg/kg至1mg/kg的剂量施用。给药可以一天一次或分成几剂进行。因此,无意以任何方式限制本发明的剂量范围。在另一方面,本公开还提供了用于杀伤肿瘤细胞的组合物,该组合物包含作为活性成分的载体,所述载体包含与特定存在于癌细胞中的核酸互补结合的多核苷酸以及编码核酸内切酶和核酸外切酶的多核苷酸。术语“癌细胞中特异性存在的核酸”、“核酸内切酶”和“核酸外切酶”如上所述。另外,“与癌细胞中特异性存在的核酸互补结合的多核苷酸”是dna。该dna核酸可以产生能够与癌细胞中特异性存在的核酸序列互补结合的crrna或grna。在此,“crrna”和“grna”如上所述。在上述组合物中,可以将i)与癌细胞中特异性存在的核酸互补结合的多核苷酸,和ii)将编码核酸内切酶的多核苷酸加载到单个载体中。如果需要,可以将i)与癌细胞中特异存在的核酸互补结合的多核苷酸,和ii)将编码核酸内切酶的多核苷酸装载在单独的载体中。另外,该组合物可以在载体中额外包含编码核酸外切酶的多核苷酸。另外,根据需要,可以将i)与癌细胞中特异性存在的核酸互补结合的多核苷酸,ii)编码内切核酸酶的多核苷酸和iii)编码核酸外切酶的多核苷酸装载在分开的载体中。在一个实施方式中,所述组合物可以是用于杀伤肿瘤细胞的组合物,其包含作为活性成分的载体,所述载体包含与癌细胞中特异存在的核酸序列互补结合的多核苷酸和编码crispr相关蛋白和核酸外切酶的融合蛋白的多核苷酸。在本公开的一个实施方式中,使用编码cas9-recj融合蛋白的多核苷酸,其中将crispr相关蛋白(核酸内切酶)cas9,与核酸外切酶recj融合,以装载在载体中。载体可以是病毒载体或质粒载体,但不限于此。可以通过本领域已知的克隆方法制备包含与癌细胞中特异性存在的核酸互补结合的多核苷酸,编码crispr相关蛋白的多核苷酸,和/或编码核酸外切酶的多核苷酸的载体,所述方法为没有特别限制。另外,一实施方式提供了治疗癌症的方法,其包括将上述用于杀伤肿瘤细胞的组合物施用于受试者。此时,与癌细胞中特异性存在的核酸互补结合的多核苷酸和核酸酶蛋白可以组合成rnp形式并施用于患有癌症的受试者。此外,可以将核酸外切酶添加到rnp中并施用于患有癌症的受试者。本公开内容的药物组合物可以通过各种途径施用于哺乳动物,例如家畜、人等。可以预期所有给药方式,例如,通过选自下组的任何一种途径进行给药,包括肿瘤内、静脉内、肌内、皮内、皮下、腹膜内、小动脉内、心室内、病灶内、鞘内、局部,及其组合。发明方式在下文中,将通过参考实施例详细描述本发明。然而,以下实施例旨在说明本发明,并且本发明的范围不仅限于此。制备实施例1:用于cas9的crrna的制备通过基因测序获得靶基因的准确核苷酸序列。在鉴定靶基因的外显子中的原间隔子相邻基序(pam,5'-ngg-3')后,确定其上游的20-mer序列为原间隔子序列。至少设计了三种原型间隔子,因为细胞中的编辑效率根据目标序列的位置而有所不同。通过将5'-tagg-3'与20-聚体序列的5'部分结合并将5'-aaac-3'与互补序列的3'部分结合来合成寡核苷酸(寡聚物)。将两种合成的低聚物调节至100μm,分别取2μl,并在46μl的纯水中稀释。使用热循环仪进行退火,将反应混合物在95℃下处理5分钟,以4℃/秒的速度冷却至55℃,然后处理10分钟。用bsai限制酶消化约5至10μg含t7启动子的用于体外转录的puc19载体(seqidno:1:taatacgactcactatagg)和crrna支架序列(seqidno:2gttttagagctagaaatagcaagttaaaataaggctagtccgttatcaacttgaaaaagtggcaccgagtcggtgc)过夜,然后纯化。其序列示为5’-taatacgactcactataggtgagaccgcaggtctcggttttagagctagaaatagcaagttaaaataaggctagtccgttatcaacttgaaaaagtggcaccgagtcggtgc-3’(seqidno:3)之后,连接用限制酶(100至200ng/l)处理后的载体。将6μl的5种退火混合物、2μl的载体、1μl的t4dna连接酶10x缓冲液(普洛麦格c126b)和1μl的t4dna连接酶(普洛麦格m180a)放入1.5mleppendorf管中,通过轻敲混合,4℃孵育过夜。用连接混合物转化大肠杆菌(e.coli)dh5α。通过sanger测序鉴定克隆,使用如下序列的m13引物:5'-taatacgactcactatagg(seqidno:1)-20聚体原间隔子序列-gttttagagctagaaatagcaagttaaaataaggctagtccgttatcaacttgaaaaagtggcaccgagtcggtgc(seqidno:2)-3’。使用由此制备的dna序列,通过聚合酶链反应(pcr)扩增包括t7启动子、原间隔子和crrna支架的区域,通过琼脂糖凝胶电泳鉴定,然后纯化。使用megashortscripttmt7试剂盒(英杰公司,am1354)在体外转录crrna,10个结果中约有800ng在50μl反应体积中反应约4到8小时。使用megacleartm试剂盒(英杰公司,am1908)纯化转录产物,并使用分光光度计测量纯化的crrna浓度。通常,获得约1-2μg/μl或更多的crrna。此时,注意防止rnase污染。将纯化的crrna稀释并以所需的浓度和体积分配,然后保存在-80℃中,同时注意避免温度变化和震动。制备实施例2:制备cpf1(cas12a)的crrna通过基因测序获得靶基因的准确核苷酸序列。在确定靶位点之后,在外显子部分中发现了一个与原间隔相邻的基序(pam,5'-tttn-3')序列,并将其下游的24-聚体序列确定为原间隔序列。此时,将原间隔序列具有约50%鸟嘌呤-胞嘧啶含量的位点确定为靶点。至少设计了三种原间隔子,因为细胞中的编辑效率根据目标序列的位置而有所不同。分别合成了包含体外转录的t7启动子和crrna的序列的寡聚体及其互补序列:5’-aattctaatacgactcactataggaatttctactgttgtagat(seqidno:25)-24聚体原间隔子序列-3'。制备最终体积为20μl的混合物,其中每种混合物含2μg合成的低聚物。此时,使用没有核酸酶的纯净水。使用热循环仪进行退火,并将反应混合物在95℃下处理5分钟,以4℃/秒的速度冷却至55℃,然后处理10分钟。使用megashortscripttmt7试剂盒(英杰公司,am1354)在体外转录crrna,5个结果中约有4μl(800ng)在50μl反应体积中反应约4到8小时。通过乙醇沉淀纯化得到的crrna,并用分光光度计测定其浓度。通常,获得约1至2μg/μl或更多的crrna,并注意防止rnase污染。将纯化的crrna稀释并以所需的浓度和体积分配,然后保存在-80℃中,同时注意避免温度变化和震动。制备实施例3:底物dna的制备使用聚合酶链反应从含有靶基因的模板中扩增出约1至1.5kbp的dsdna,以使cas9或cpf1的原间隔子靶向的核苷酸序列位于中间区域。在此,原间隔子是指grna可以互补结合的宿主细胞的dna中的靶核苷酸序列。通过克隆将dsdna插入puc19或pgem载体后,使用m13引物进行测序以确认原间隔子序列。使用m13引物通过聚合酶链式反应扩增底物dna,纯化,并以100ng/μl的浓度保存在-20℃中。实施例1:证实crrna激活crispr/cas蛋白的非特异性核酸酶功能研究证实,包括crispr/cas12a在内的crispr/cas蛋白的基因组编辑功能被crrna引导的靶序列结合激活,从而激活了降解dna或rna分子的非特异性核酸酶功能。其示意图如图1所示。实施例2:通过crisprplus识别癌细胞特异性snp和癌细胞凋亡根据癌症起源的组织和癌症的类型,癌细胞具有其自身的染色体突变,包括正常细胞中不存在的特定基因组区域中的基因突变或单核苷酸多态性(snp)。这些癌症特异性snp被用作本发明的癌细胞特异性标记。癌细胞特异性snp用于合成含有与snp互补序列的crrna,并被含有crrna和核酸外切酶的crispr相关蛋白的crisprplus蛋白识别。癌细胞基因组中snp和crrna之间的序列特异性结合激活了crisprplus蛋白的基因组编辑功能,导致靶标dna/rna断裂。上述激活随后激活了crisprplus的固有非特异性核酸酶功能,该功能不可逆地破坏了癌细胞中的ds和ssdna/rna分子,从而导致凋亡。这些结果在图2中以示意图示出。实施例3:spycas9(seqidno:46)的核酸内切酶功能验证(体外)将nebuffertm3.1在无核酸酶的纯水中稀释至最终的1x浓度,并向其中添加120nm的核酸酶和120nm的crrna,诱导rnp复合物形成。混合后,将混合物在室温下孵育约15分钟。向其中添加约200ng的底物dna,轻敲混合物,在37℃下反应。为了确定与没有靶标序列的底物dna的反应,在这一步中加入不能被靶向的dna。将反应混合物的最终体积调节至20μl。反应后,加入负载凝胶的染料溶液并充分混合。制成2%琼脂糖凝胶(琼脂糖,sepro,gendepot,a0224-050)后,使用1kbdna标记(赛默科技,sm0311),电泳12μl的染色反应物。随后,观察到核酸酶活性所致的底物dna条带切割。结果证实,当只有无靶序列的底物dna被反应时,dna不被切割。然而,已经证实,当将靶标dna放在一起时,所有dna都被切割。实施例4:cpf1的非特异性核酸外切酶功能的证实(体外)将nebuffertm1.1在无核酸酶的纯水中稀释至最终的1×浓度,并向其中加入120nm的crispr/cas12a和120nm的crrnadhcr7,诱导rnp复合物形成。向230nm的rnp复合物中,小心地加入200ng的底物dna,轻敲混合物,在37℃下反应。此时,由将单独的特异性或非特异性dna底物,或特异性和非特异性dna的混合物在nebuffer1.1buffer中,37℃条件下孵育1.5小时或24小时来制备底物dna。将反应混合物的最终体积调节至20μl。在反应所需时间后,加入载有凝胶的染料并充分混合。制成2%琼脂糖凝胶(琼脂糖,sepro,gendepot,a0224-050)后,使用1kbdna标记(赛默科技,sm0311),电泳12μl的染色反应物。观察到核酸酶活性所致的底物dna条带切割。结果如图3所示。为了证明crispr核酸酶具有序列非特异性核酸外切酶活性,使用crispr核酸酶、特异性和非特异性dna底物进行体外dna切割实验,结果表明,crispr核酸酶的非特异性核酸外切酶活性取决于关于序列特异性核酸内切酶的活性。具体地,将crispr/cas12a,靶向人dhcr7基因的crrna,和具有crrna靶向序列的特定dna底物(dna#1,1.5kb),或不具有crrna靶向序列的非特异性dna底物(dna#2,0.5kb)孵育1.5或24小时以诱导dna切割,通过琼脂糖凝胶进行验证(见图3)。此处,与dhcr7、dsdna1(1,431bp)和dsdna2(544bp)互补的crrna序列分别由seqidno:132、seqidno:133,以及seqidno:134表示。当特异性dna底物与核酸酶和crrna一起孵育1.5小时时,底物被序列特异性地切割成约0.7kb的片段(上图,泳道3)。然而,实验证实,没有crrna的情况下底物不被切割(上图,泳道4)。当在相同条件下用非特异性dna进行孵育时,无论crrna是否存在,dna切割都不会发生(上图,泳道5和6)。当特异性dna底物和非特异性dna底物同时用核酸酶处理时,仅特异性dna底物如预期被切割(上图,泳道7和8)。另外,当特异性dna底物、核酸酶和crrna的孵育时间增加至24小时时,观察到特异性dna底物及其片段消失(下图,泳道3)。这意味着该dna被crispr核酸酶的核酸外切酶活性切割。当在不存在crrna的情况下进行相同实验时,dna没有消失(下图,泳道4),表明核酸外切酶活性取决于crispr/cas12a的序列特异性酶活性。此外,结果还证明了这样的事实:当用核酸酶和crrna处理非特异性dna24小时(下图,泳道5和6)时,dna被保留而没有消失。此外,当同时用核酸酶和crrna处理特异性和非特异性dna底物24小时时,特异性和非特异性dna底物均被降解并消失(下图,泳道7),只有在crrna存在的情况下被观测到(下图,泳道8)。这意味着通过激活序列特异性核酸内切酶功能诱导的crispr/cas12a核酸外切酶功能以序列非特异性方式起作用。因此,这些实验结果暗示crispr/cas12a具有取决于序列特异性酶活性的非特异性核酸外切酶活性。实施例5:细胞毒性分析在5%co2培养箱中,使用dmem/10%fbs生长培养基在37℃中培养源自人类癌症的细胞,即hela细胞。转染前一天,将2.5×104细胞悬浮在100μl培养基中,在96孔板中铺板。空白(背景对照)孔仅加入100μl培养基。次日,在下表3所示的条件下用crispr/cas核酸酶和crrna的复合物(rnp)转染。其中一个crrna对人dhcr7基因具有序列特异性,另一个对水稻dwarf5基因具有序列特异性。[表3]对于每个孔,将5μl的opti-mem培养基,2.4nmcrispr/cas和2.4nmcrrna,在1.5ml试管中混合,在室温下孵育10分钟。将0.17μl的脂质转染胺casplus试剂加入到同一试管中,并在室温下孵育5分钟。在上述管的孵育过程中准备另一管。将5μlopti-mem和0.3μl脂质转染胺crisprmax试剂在试管中混合,然后在室温下孵育5分钟。将两个试管的内容物混合并在室温下孵育10分钟。将得到的试管溶液逐滴添加到细胞生长的每个孔中。然后将细胞在5%co2培养箱中于37℃进行培养。在24、48和72小时后,在清洁工作台上,将10μlwst-1(细胞增殖试剂,罗氏0501594401)添加到每个孔中。然后,将板置于温度为37℃的5%co2培养箱中,观察到颜色变化(浅红色→深红色)。十分钟后,使用fluostaromegaelisa读数器(bmglabtech)在420至480nm和690nm处测量背景和样品的吸光度。分析crispr/cas核酸酶在靶向和非靶向crrna中的细胞毒性。将源自人类的癌细胞,hek293(图4a)和hela(图4b)分别用crispr/cas12a核酸酶、crrna或这两种分子的结合物(rnp复合体)转染,然后在24、48、72小时后,通过使用wst-1的活力测定法测量细胞活力。结果表明,用核酸酶或crrna转染的细胞仅在24、48和72小时时,显示存活率基本无变化(见图4)。这些结果表明核酸酶和crrna本身对细胞没有毒性。另一方面,用具有序列特异性酶活性的结合物转染的细胞在72小时时,活力显著降低。该结果表明crispr/cas12a核酸酶对细胞表现出的毒性取决于序列特异性酶的活性。用相同结合物转染的细胞在24和48小时后表现为活力未变化(hek293)或活力略有下降(hela)。这意味着crispr/cas12a核酸酶的毒性影响细胞需要一些时间。一般地,已知crispr/cas核酸酶的序列特异性酶活性在细胞中从24小时到48小时是稳定存在的。此外,鉴于该活性导致非特异性核酸酶活性表现出细胞毒性,可以说72小时后出现的细胞毒性并不是独立于核酸酶活性的间接效应,而是由核酸酶的序列特异性酶活性相关功能引起的。然而,已证实使用特异性crrna比使用非特异性crrna更具细胞毒性。因此,该实验的结果表明,通过依赖于序列特异性酶活性而对细胞表现出毒性,crispr/cas12a核酸酶具有降低细胞活力的功能。实施例6:癌细胞特异性毒性分析通过分析人源性肺癌细胞和正常细胞的基因序列,发现了癌细胞中特异性存在snp,并合成了能够靶向它们的crrna。将crispr核酸酶和crrna混合以制备rnp复合物,然后用其转染癌细胞和正常细胞。采用基于wst-1的细胞活力测定法分析了癌细胞特异性杀伤效果。制备使用过的crrna,以靶向特异性存在于肺癌的egfr中的序列(seqidno:43)。转染前一天,将2.5×104细胞悬浮在100μl培养基中,在96孔板中铺板。空白(背景对照)孔仅加入100μl培养基。次日,在下表4所示的条件下用crispr/cas核酸酶和crrna的复合物(rnp)转染。[表4]对于每个孔,将5μl的opti-mem培养基,2.4nmcrispr/cas和2.4nmcrrna在1.5ml试管中混合,在室温下孵育10分钟。将0.17μl的脂质转染胺casplus试剂加入到同一试管中,并在室温下孵育5分钟。在上述管的孵育过程中准备另一管。将5μlopti-mem和0.3μl脂质转染胺crisprmax试剂在试管中混合,然后在室温下孵育5分钟。将两个试管的内容物混合并在室温下孵育10分钟。将得到的试管溶液逐滴添加到细胞生长的每个孔中。然后将细胞在5%co2培养箱中于37℃进行培养。在24、48和72小时后,在清洁工作台上,将10μlwst-1(细胞增殖试剂,罗氏0501594401)添加到每个孔中。然后,将板置于温度为37℃的5%co2培养箱中,观察到颜色变化(浅红色→深红色)。十分钟后,使用fluostaromegaelisa读数器在420至480nm和690nm处测量背景和样品的吸光度。分析crispr/cas核酸酶在靶向和非靶向crrna中的细胞毒性。结果证实,在仅使用靶向crrna处理的组中特异性杀死了肺癌细胞。实施例7:验证egfr突变体序列特异性引导rna所致的凋亡作用在该实施例中,已经证明使用cas9蛋白表达载体(px459,addgene质粒#62988)可以在hcc827肺癌细胞的基因组中引起多重切割,从而诱导细胞凋亡。通过电穿孔将载体转导到细胞中。为了诱导基因组的多重切割,使用了靶向hcc827细胞中egfr基因e2突变序列的向导rna。已知egfr基因的e2突变序列存在于18个以上的多份拷贝中。将hcc827细胞在含有rpmi-1640(10%胎牛血清)培养基的75t烧瓶中培养至约50%的融合度,胰蛋白酶消化,pbs洗涤,最后在neon电穿孔缓冲液r中重悬。将150,000个细胞和500ng载体加载至10μlneon移液器吸头中,在1300v,20ms和2个脉冲的条件下进行电穿孔。然后,细胞在rpmi-1640(10%胎牛血清)培养基中恢复,在6天后收获并计数。结果示于图5和6中。如图5和图6所示,结果证实,与电脉冲组相比,没有靶序列的egfr_wt实验组(pspcas9(bb)-2a-puro(px459)v2.0-egfr_wt)相比egfr_e2实验组(pspcas9(bb)-2a-puro(px459)v2.0-egfr_e2),细胞数明显减少。具体而言,在egfr_e2实验组中诱导了83%的细胞凋亡。这些结果证实,向癌细胞中添加cas9蛋白和细胞遗传序列特异性多靶标向导rna诱导了癌细胞的凋亡。实施例8:验证肺癌细胞h1299中依赖于靶位的凋亡作用实施例8.1:通过脂质转染引入grna和crispr相关蛋白将肺癌细胞h1299以1.5×105细胞/孔的量接种在24孔板中。24小时后,将下表5所示的dna种类(ccr5、hprt1、mt2、smim11、gnpda2、slc15a5和kcne2)引入每个孔中。为了进行引入,根据制造商的说明书,分别对每500ngdna使用脂质转染胺3000试剂。[表5]从dna导入之时,即转染的72小时后,用抽吸法除去各孔的培养液,并用500μl的1×pbs洗涤细胞一次。然后,将胰蛋白酶-edta加入孔中以分离所有细胞。用台盼蓝染料将细胞染色,并计数活细胞的数目。结果示于图7和8中。如图8所示,nt(非靶标),即在h1299中没有可切割的靶序列,而ccr5和hprt1有一对靶位位点,表现出类似的凋亡作用。另一方面,mt2,已知可以切割100多个位点,通过将活细胞的数量减少到约50%,显示出一种凋亡效应。此外,肺癌细胞株的靶位smim11(约74%)、gnpda2(约58%)、slc15a5(约45%)和kcne2(约77%)显示出与mt2一样高的凋亡效应。其中,smim11和kcne2显示出比mt2更好的凋亡效应。每个实验组的拷贝数和重要性示于下表6,在显微镜下观察各组细胞形态,示于图7中。[表6]实施例8.2:使用脂质转染胺引入grna和crispr相关蛋白将h1299的肺癌细胞以1.5×105细胞/孔的量接种在24孔板中。24小时后,将下表7所示种类的dna引入每个孔中。为了进行引入,根据制造商的说明书,分别对每500ngdna使用脂质转染胺3000试剂。[表7]导入dna,即转染48小时后,用抽吸法除去各孔的培养液,并用500μl的1×pbs洗涤细胞一次。然后,将胰蛋白酶-edta加入孔中以分离所有细胞。用台盼蓝染料将细胞染色,并计数活细胞的数目。结果示于图9和10中。在该实施例中,包括其中仅进行脂质转染而不导入dna的实验组。如图9所示,与仅脂质转染(仅lipo)相比,ccr5中的活细胞数量减少至约80%。与ccr5相比,nt1和hprt1约为70%。与nt1相比,mt2中的细胞数减少到约25%。在靶向cnv的三种细胞(gnpda2、slc15a5和kcne2)中,更多的细胞死亡,台盼蓝细胞计数无法显著检测活细胞。特别地,在nt1、ccr5、hprt1、mt2、gnpda2、slc15a5和kcne2中显示出约43%、约23%、约50%、约86%、约99%、约99%和约99%的细胞凋亡率。分别选择nt1和gnpda2实验组作为具有大量活细胞的实验组和具有大量死细胞的实验组的代表。之后,通过nucbluelivereadyprobes试剂和碘化丙啶readyprobes试剂在显微镜下对每个孔成像。结果如图10所示。通过荧光分析活细胞的比例,结果与图9所示相似。实施例9:验证肺癌细胞h1563中依赖于靶位的凋亡作用将h1563细胞以1.5×105细胞/孔接种在24孔板中,并且在24小时后,将下表8中所示种类的dna导入每个孔中。为了进行导入,根据制造商的说明书,分别对每500ngdna使用脂质转染胺3000试剂。[表8]从导入dna之时,即转染后24小时,将嘌呤霉素以1μg/ml的浓度加入到每个孔中,并筛选72小时。然后,通过抽吸除去各孔的培养液,并用500μl的1xpbs洗涤细胞一次。此后,将胰蛋白酶-edta应用于孔以分离所有细胞。用台盼蓝染料将细胞染色,并计数活细胞的数目。结果如图11所示。如图11所示,相比于单一靶标ccr5,有少量的细胞保留,分别为hprt1中约52%,mt2中约43%,两种cnv靶标(irx1和adamts16)中约37%和40%。从这些结果可以确认,与ccr5等单个靶标相比,通过诱导大量的双链断裂剂(dsb),可以提高凋亡作用。实施例10:验证肺癌细胞a549中依赖于靶位的凋亡作用实施例10.1:多靶点验证a549细胞凋亡通过电穿孔(lonza)将500ng表达cas9蛋白和grna的dna导入肺癌细胞系a549的细胞中。用1×pbs洗涤培养物中的a549细胞,然后用胰蛋白酶-edta处理以使其从底部脱离。取出所需数量的细胞,用1×pbs洗涤一次,悬浮在sf缓冲液(lonza)中,然后与每种dna混合。将细胞和dna的混合物置于电穿孔器(lonza)中并施加电击。作为对照,以在人类基因序列中不比对的nt1为靶标的条件;hprt1,一个单拷贝的管家基因;ccr5,单拷贝的基因和仅电击(仅使用脉冲)条件。通过电击导入dna后,将细胞接种于24孔板中,一式两份,接种于96孔板中,一式三份。从导入dna开始24、42和72小时后,将50μlcelltiterglo试剂添加到96孔板的每个孔中。将板置于fluostaromega读取器上并振动2分钟。使它们在室温下反应10分钟后,测量荧光。上述方法是一种通过发光程度,根据细胞内atp的含量来确定正在进行代谢的活细胞数量的方法。结果如图12所示。如图12所示,与三个对照(nt1、hprt1和ccr5)相比,在靶向100个或更多位点的mt2条件下,随着时间的推移诱导了20%至50%的凋亡。导入dna后24小时,将1μg/ml的嘌呤霉素添加到24孔板的每个孔中。在仅电击的情况下,细胞凋亡约有90%时,更换细胞培养基使细胞恢复。恢复培养5至7天后,将细胞从每个孔中分离并用台盼蓝染色,并计数活细胞的数目。结果如图13所示。另一方面,图14显示了在计数细胞数之前,dna计数和使用嘌呤霉素筛选后,用显微镜观察到的照片。结果显示,细胞在仅电击而没有嘌呤霉素抗性的条件下不能存活,成为对照,当靶向ccr5时,与nt1和hprt1的条件相比,观察到约50%的细胞凋亡。并且证实了大约90%的细胞在mt2条件下死亡。导入dna后,用嘌呤霉素选择细胞,然后在计数细胞数之前在显微镜下观察。与细胞计数的结果相似,与nt1对照相比,靶向hprt1和ccr5(仅具有一个拷贝)时,超过50%的细胞得以恢复。另一方面,在mt2条件下,约90%的细胞被杀死并且仅10%的细胞恢复(蓝色箭头:恢复的细胞集落)。因此,证实了当使用cas9蛋白在a549细胞中诱导多dna断裂时诱导了凋亡。实施例10.2:验证cnv靶标所致的a549细胞凋亡在肺癌细胞系a549的细胞中,通过靶向含有cnv的基因,用cas9蛋白诱导dna断裂,并检测凋亡程度。具体地,通过电穿孔(lonza)将500ng表达cas9蛋白的dna和每个靶向cnv的grna引入a549细胞。用1×pbs洗涤培养物中的a549细胞,然后用胰蛋白酶-edta处理以使其从底部脱离。取出所需数量的细胞,用1×pbs洗涤一次,悬浮在sf缓冲液(lonza)中,然后与每种dna混合。将细胞和dna的混合物置于电穿孔器(lonza)中并施加电击。作为对照,添加了导入pet21a载体在大肠杆菌表达蛋白质的条件,仅电击条件(仅脉冲)和不使用处理(无脉冲)条件。通过电击导入dna后,将细胞按条件分别铺板在96孔板中,一式三份。dna导入24、44和51小时后,将50μlcelltiterglo试剂添加到每个孔中。将板置于fluostaromega读取器上并摇动2分钟。使它们在室温下反应10分钟后,测量荧光。结果如图15所示。如图15所示,与三个对照(仅脉冲、pet21a和无脉冲)相比,在靶向100个或更多位点的mt2条件下,随着时间的推移诱导了20%至50%的细胞凋亡。此外,与对照组相比,cd68、dach2和herc2p2的三个cnv靶标可诱导mt2类似的凋亡,而shbg的cnv靶标可诱导70%至80%的凋亡。因此,证实了当使用靶向a549细胞中的四种cnv(cd68、dach2、herc2p2和shbg)的cas9蛋白诱导靶dna断裂时,诱导了凋亡。实施例11:验证乳腺癌细胞skbr3中依赖于靶位的凋亡作用实施例11.1:cnv靶标验证skbr3细胞凋亡通过电穿孔(lonza)将500ng表达cas9蛋白的dna和每个靶向cnv的grna导入乳腺癌细胞系skbr3的细胞中。用1×pbs洗涤培养物中的skbr3细胞,然后用胰蛋白酶-edta处理以使其从底部脱离。取出所需数量的细胞,用1×pbs洗涤一次,悬浮在sf缓冲液(lonza)中,然后与每种dna混合。将细胞和dna的混合物置于电穿孔器(lonza)中并施加电击。作为对照,添加了在人类基因序列中未比对的靶标nt1的条件(非靶标),仅电击的条件(仅脉冲)和不处理的条件(无脉冲)。通过电击导入dna后,将细胞放入24孔板中,重复两次,放入96孔板中,重复三次。在导入dna24、42和48小时后,将50μlcelltiterglo试剂添加到96孔板的每个孔中。将板置于fluostaromega读取器上并振动2分钟。使它们在室温下反应10分钟后,测量荧光。结果如图16所示。如图16所示,与三个对照(nt1、脉冲,无脉冲)相比,在靶向100个或更多位点的mt2条件下,随着时间的推移诱导了30%的细胞凋亡。此外,与对照相比,erbb2和krt16的两个cnv靶标诱导了40%至50%的凋亡。实施例11.2:验证cnv靶标所致的skbr3细胞凋亡通过电穿孔(lonza)将500ng表达cas9蛋白的dna和每个靶向cnv的grna导入乳腺癌细胞系skbr3的细胞中。用1×pbs洗涤培养物中的skbr3细胞,然后用胰蛋白酶-edta处理以使其从底部脱离。取出所需数量的细胞,用1×pbs洗涤一次,悬浮在sf缓冲液(lonza)中,然后与每种dna混合。将细胞和dna的混合物置于电穿孔器(lonza)中并施加电击。作为对照,使用以人基因序列中不一致的nt1和单拷贝的管家基因hprt1为靶点。通过电击导入dna后,将细胞放入24孔板中,重复两次。导入dna后48小时,将细胞与24孔板的每个孔分离,并用台盼蓝染色,并计数活细胞的数目。结果如图17所示。如图17所示,与对照组相比,在靶向100个或更多位点的mt2条件下,随着时间的推移,诱导了40%的细胞凋亡,与对照组相比,erbb2和krt16的两个cnv靶标诱导了40%至50%的细胞凋亡。因此,证实了当使用靶向skbr3细胞中的两种cnv(erbb2和krt16)的cas9蛋白诱导靶dna断裂时,诱导了凋亡。实施例12:验证宫颈癌细胞hela中依赖于靶位的凋亡作用实施例12.1:通过cnv靶标和hpv基因靶标确定凋亡通过电穿孔将600ng表达cas9蛋白和grna的载体导入宫颈癌细胞系hela细胞。用1×pbs洗涤培养的hela细胞,然后用胰蛋白酶-edta处理以使其从底部脱离。取出所需数量的细胞,用1×pbs洗涤一次,悬浮在sf缓冲液(lonza)中,然后与每种载体混合。将细胞和dna载体的混合物置于电转器(lonza)中并施加电击。作为对照,使用靶向2个拷贝的非必需基因ccr5的条件和靶向超过100个非必需基因mt2条件。对实验组进行实验,通过靶向已知存在有8个或更多拷贝的prdm9,和靶向已知存在于hela细胞中30个拷贝或更多的人类乳头瘤病毒(hpv)衍生的基因来诱导hela细胞特异性凋亡。通过电击导入dna后,将细胞放入24孔板中,重复两次。24小时后,向每个孔中加入0.5μg/ml的嘌呤霉素进行筛选,以杀死没有dna载体的细胞。筛选3天后,将培养基更换为不含嘌呤霉素的培养基进行培养。3至4天后,将细胞从每个孔中分离并用台盼蓝染色,并计数活细胞的数目。结果示于图18和19中。如图18和19所示,已经证实,与仅在hela细胞中的切割一个位点的ccr5相比,随着时间的推移,约50%的细胞凋亡是由靶向100个或更多位点的mt2条件诱导的。此外,研究证实,cnv靶标prdm9和hpv基因靶标hpv_1诱导的细胞凋亡与mt2条件相似或更多。特别地,在图18中,mt2和prdm9分别显示出约50%和约80%的凋亡率。另外,在图19中,在mt2、prdm9和hpv_1中观察到约50%、约40%和约40%的细胞凋亡率。因此,实验证实,当使用仅存在于hela细胞中的cnv和hpv基因的cas9蛋白诱导靶标dna断裂时,诱导凋亡。实施例12.2:验证cnv靶标和hpv基因靶标所致的凋亡通过电击将dna载体导入hela细胞的过程与实施例12.1相同。通过电击导入dna后,将细胞放入96孔板中,重复两次。此后的24、48和72小时,使用celltiterglo2.0测量细胞中atp的量,以通过发光程度确定进行代谢过程的活细胞的量。作为对照,添加了表达gfp的对照载体,使用靶向1个拷贝的非必需基因ccr5的条件,添加靶向超过100个非必需基因mt2条件。对实验组进行实验,通过靶向已知存在有8个或更多拷贝的prdm9,和靶向已知存在于hela细胞中30个拷贝或更多的人类乳头瘤病毒(hpv)衍生的基因来诱导hela细胞特异性凋亡。结果显示在图20a中。如图20a所示,仅切割hela细胞中一个位点的ccr5靶标和包含gfp的仅电击对照显示了相似的发光信号。此外,证实了与ccr5条件相比,mt2条件诱导了约50%的凋亡。此外,证实了cnv靶标prdm9和hpv基因靶标hpv_1比mt2条件更多地诱导凋亡。具体地,mt2、prdm9和hpv_1分别显示出约50%、约65%和约65%的凋亡率。因此,实验证实,当使用仅存在于hela细胞中的cnv和hpv基因的cas9蛋白诱导靶标dna断裂时,诱导凋亡。实施例12.3:验证hpv基因靶标所致的凋亡通过电击将dna载体导入hela细胞的过程与实施例12.1相同。在靶标1个拷贝的非必需基因ccr5的现有条件和靶标100个以上非必需基因的mt2条件中,添加了靶向人类基因组中不存在区段的nt条件。nt1是表达20-mer非靶标sgrna的条件,nt2和nt3是非靶标sgrna的间隔子长度分别为10mer和5mer的条件。对实验组进行实验,通过靶向人乳头瘤病毒(hpv)衍生的基因来诱导hela细胞特异的凋亡,已知该基因在hela细胞中存在30拷贝或更多。通过电击导入dna后,将细胞放入24孔板中,重复两次。24小时后,向每个孔中加入0.5μg/ml的嘌呤霉素进行筛选,以杀死没有dna载体的细胞。筛选3天后,将培养基更换为不含嘌呤霉素的培养基进行培养。10天后,将细胞从每个孔中分离,并通过celltiterglo方法测量发光信号。结果显示在图20b中。如图20b所示,与具有5-mer间隔子的nt3条件相比,仅切开hela细胞中一个位点的ccr5靶标显示了约75%的发光信号,与nt3条件相比,mt2条件下诱导了约99.5%的凋亡。此外,证实了与ccr5条件相比,mt2条件诱导了约50%的凋亡。另外,与nt3条件相比,hpv基因的靶标hpv_1可杀死约90%的细胞。因此,通过celltiterglo方法证实了当使用仅存在于hela细胞中的hpv基因的cas9蛋白诱导靶dna断裂时,诱导凋亡。实施例13:验证结肠癌细胞ht-29中依赖于靶位的凋亡作用实施例13.1:验证cnv靶标所致的凋亡将ht-29细胞以1.5×105细胞/孔接种在24孔板中。24小时后,将下表9所示种类的dna引入每个孔中。为了进行导入,根据制造商的说明书,分别对每500ngdna使用脂质体3000试剂。[表9]从dna导入(即转染)起24小时后,将嘌呤霉素以1μg/ml的浓度加入各孔中,并筛选90小时。然后,通过抽吸除去每个孔的培养液,并使细胞在正常培养基(mccoy+10%fbs,1%p/s)中恢复12天。然后,将细胞用500μl/孔的1×pbs洗涤一次,并将胰蛋白酶-edta加入孔中以分离所有细胞。用台盼蓝染料对细胞染色,将活细胞数计数两次,然后取平均值。结果如图21所示。如图21所示,已知没有靶序列的nt1和具有单个靶标的ccr5显示出在误差范围内的差异。对于hprt1,与nt1相比,约47%的细胞存活。mt2是一种阳性对照,可切割整个基因组中的100个重复序列,并且与nt1相比,四个cnv靶标(trappc9、linc00536、trps1和cdk8)的细胞存活率分别为2%、3%、18%、20%和4.2%。即实验证实,通过使用cnv靶可以有效地杀伤癌细胞。实施例13.2:验证cnv靶标所致的结肠癌细胞凋亡为了鉴定通过靶向cnv(cdk8、linc00536、trps1和trappc9)和mt2的四个基因所致的ht-29(结肠癌细胞系)的特异性细胞凋亡,将500ng表达cas9蛋白的dna和每个靶向cnv的grna通过电穿孔(lonza)导入ht-29细胞。用1×pbs洗涤培养物中的ht-29细胞,然后用胰蛋白酶-edta处理以使其从底部脱离。取出所需数量的细胞,用1×pbs洗涤一次,悬浮在sf缓冲液(lonza)中,然后与每种dna混合。将细胞和dna的混合物置于电穿孔器(lonza)中并施加电击。作为对照,添加了在人类基因组中未对齐的靶标nt1的条件(非靶标)和仅电击的条件(仅脉冲)。电击后,将细胞加入96孔板中,每个条件重复四次。dna导入24小时后,将50μlcelltiterglo试剂添加到每个孔中。将板置于fluostaromega读取器上并摇动2分钟。在室温下反应10分钟后,测量发光。结果如图22所示。如图22所示,与对照组相比,在靶向100个或更多位点的mt2条件和靶标trappc9cnv条件下诱导了90%的凋亡,而三个靶标cdk8、linc00536和trps1的cnv则诱导了20%至45%的凋亡。实施例14:比较肺癌细胞h1299与h1563中所用的靶基因的凋亡作用将h1563细胞以1.5×105细胞/孔接种在24孔板中。24小时后,将下表10所示种类的dna引入每个孔中。为了进行导入,根据制造商的说明书,分别对每500ngdna使用脂质体3000试剂。[表10]条件ccr5smim11gnpda2slc15a5kcne2切割数22222必需性非-必需非-必需癌基因癌基因非-必需导入dna24小时后,即转染后,将嘌呤霉素以1μg/ml的浓度加入到每个孔中,并筛选72小时。然后,通过抽吸除去每个孔的培养液,将细胞用500μl/孔的1×pbs洗涤一次,并将胰蛋白酶-edta应用于孔以分离所有细胞。用台盼蓝染料将细胞染色,并计数活细胞的数目。结果如图23所示。此时,除ccr5外,所有使用的dna靶均在h1299中扩增,并通过切割基因组上的12个以上位点而显示出凋亡作用(参见图8和图9)。但是,由于在h1563细胞中不存在这种扩增的cnv,因此与ccr5一样,所有靶标仅切割两个靶标位置。如图23所示,在肺癌细胞h1563中,这些靶标未像h1299肺癌细胞那样显示出凋亡作用,但与ccr5相比显示出更高的细胞存活率。但是,在kcne2的情况下,在h1563中也观察到了高凋亡作用。这与从图24的显微图像中推断出的细胞凋亡趋势一致。推测kcne2的凋亡作用是出于未知原因发生的,并且h1299的三个cnv靶标(smim11、gnpda2和slc15a5)(除kcne2之外)均未诱导h1563的凋亡。因此,证实了cnv的细胞特异性凋亡作用。实施例15:检测cnv靶标所致的肺癌h1299细胞凋亡转染前一天,用胰蛋白酶-edta分离肺癌细胞系h1299的细胞,并以1.3×104细胞/孔的量接种在白色的96孔板中。第二天,通过脂质体的方法将500ng表达cas9蛋白的dna和每个靶向cnv的grna引入细胞。混合0.3μl脂质胺试剂i和5μlopti-mem(试管1)。将每种条件下的0.2μl脂质体试剂ii,5μlopti-mem和500ngdna混合以制备试管2。混合两个管的内容物,室温放置15分钟。将脂质体和dna的混合物以11μl/孔添加到96孔板的孔中。3小时后,将annv试剂加入孔中,并在转染24小时后测量发光度。结果如图25所示。该方法是根据当凋亡时annv附着到暴露于细胞外膜的ps(磷脂酰丝氨酸)位点时产生的发光程度来测量凋亡程度的方法。实验结果表明,kcne2、gnpda2、smim11和slc15a5的凋亡率约为30%至40%。根据以上结果,当使用cas9蛋白诱导h1299细胞的四个cnv(gnpda2、kcne2、slc15a5和smim11)中的靶标dna断裂时,证实了凋亡是由于ps暴露于细胞膜引起的。实施例16:确认egfr突变序列特异性向导rna所致的凋亡作用以及使用cas9-recj融合蛋白(crisprplus)的凋亡作用在该实施例中,已经证明使用cas9蛋白表达载体(px459,addgene质粒#62988)可以在hcc827肺癌细胞的基因组中引起多重切割,从而诱导细胞凋亡。此外,证实了通过px459载体与cas9蛋白一起表达人密码子优化的recj蛋白可以增强凋亡作用。使用电穿孔将载体转移到细胞中,使用靶向hcc827细胞中egfr基因的e2突变序列的向导rna以便诱导基因组的多重切割。将hcc827细胞在含有rpmi-1640(10%胎牛血清)培养基的75t烧瓶中培养至约50%的融合度,胰蛋白酶消化,pbs洗涤,最后在neon电转化缓冲液r中重悬。将150,000个细胞和500ng载体加载至10μlneon移液器吸头中,在1300v,20ms和2个脉冲的条件下进行电穿孔。然后,细胞在rpmi-1640(10%胎牛血清)培养基中恢复,在4天后收获并计数。结果示于图26和27中。参照图26和27,与仅施加电脉冲(仅脉冲)的对照组相比,与没有靶序列的egfr_wt实验组相比,egfr_e2实验组中的细胞数目显著减少。具体地,使用cas9靶向ccr5的实验组的细胞数没有变化,但是靶向egfr_e2的实验组的细胞凋亡率约为33%。此外,实验观察到,当recj蛋白被共表达,单个靶标被切割时,不仅通过多切割放大了凋亡作用,而且减少了细胞数目。具体地,使用cas9-recj靶向ccr5的实验组显示约50%的凋亡率,而使用cas9-recj靶向egfr_e2的实验组显示约80%的凋亡率。通过上述实验,发现将cas9蛋白和细胞基因组序列特异性多靶标引导rna引入癌细胞时,可以诱导细胞凋亡,并且可以通过crisprplus蛋白来控制其作用。实施例17:确认egfr突变序列特异性的向导rna所致的凋亡作用以及使用rnp的凋亡作用在该实施例中,已经证明使用cas9/sgrna核糖核蛋白(cas9rnp)可以在hcc827肺癌细胞的基因组中引起多重切割,从而诱导凋亡。通过电穿孔将rnp转导到细胞中,通过使用靶向hcc827细胞中egfr基因的e2突变序列的向导rna以诱导基因组的多重切割。将hcc827细胞在含有rpmi-1640(10%胎牛血清)培养基的75t烧瓶中培养至约50%的融合度,胰蛋白酶消化,pbs洗涤,最后在neon电转化缓冲液r中重悬。将150,000个细胞和1.2μm的rnp加入10μlneon移液器吸头中,在1300v,20ms和2个脉冲的条件下进行电穿孔。然后,使细胞在rpmi-1640(10%胎牛血清)培养基中恢复培养,在1天后收获并计数。结果如图28所示。如图28所示,与仅施加电脉冲的对照组(仅脉冲)或加入cas蛋白或仅使用向导rna的对照组相比,引入靶向egfr_e2序列的rnp的实验组中的细胞数量明显减少。具体地,引入靶向egfr_e2序列的rnp的实验组显示出约33%的细胞凋亡率。通过以上实验,证实了当将cas9蛋白和细胞基因组序列特异性多靶标向导rna导入癌细胞时,可以诱导细胞凋亡,并且可以通过cas9rnp来控制其作用。实施例18:确认mt2序列特异性的向导rna所致的凋亡作用以及使用rnp的凋亡作用在该实施例中,已经证明使用cas9/sgrna核糖核蛋白(cas9rnp)可以在h1563肺癌细胞的基因组中引起多重切割,从而诱导凋亡。电穿孔被用于将rnp转移到细胞中,通过在人类基因组中的靶向100多个位点的靶向mt2的向导rna,以诱导基因组的多重切割。将h1563细胞在含有rpmi-1640(10%胎牛血清)培养基的75t烧瓶中培养至约50%的融合度,胰蛋白酶消化,pbs洗涤,最后在neon电转化缓冲液r中重悬。将150,000个细胞和1.2μm的rnp加入10μlneon移液器吸头中,在1,200v,20ms和2个脉冲的条件下进行电穿孔。然后,细胞在rpmi-1640(10%胎牛血清)培养基中恢复,在2天后收获并计数。结果如图29所示。如图29所示,证实了与仅施加电脉冲的对照组(仅脉冲)或与仅引入向导rna的对照组相比,引入靶向mt2序列的rnp的实验组中的细胞数量明显减少。具体地,引入了靶向mt2序列的rnp的实验组表现出约35%的细胞凋亡率。通过以上实验,证实了当将cas9蛋白和细胞基因组序列特异性多靶标引导rna引入癌细胞中时,诱导了细胞凋亡并且可以通过rnp控制该作用。实施例19:确认mt2、gnpda2和smim11序列特异性的向导rna所致的凋亡作用以及使用rnp的凋亡作用在该实施例中,已经证明使用cas9蛋白可以在作为肺癌细胞的h1299细胞的基因组中引起多重切割,从而诱导细胞凋亡。在各种癌细胞中,实验中使用具有较高转染效率的h1299细胞,通过电穿孔将rnp转移到细胞中。作为诱导基因组多重切割的向导rna,使用了靶向以下基因的三种向导rna:mt2、gnpda2和smim11。基于h1299细胞特异性基因组测序信息,制备分别靶向约12个或更多拷贝存在的癌基因(gnpda2)和以40个或更多拷贝存在的非必需基因(smim11),高拷贝数变化(cnv)的向导rna。分析了人类基因组序列中存在的所有cas9原间隔相邻基序(pam,5'-ngg-3'),并构建了靶向mt2的向导rna,该rna可以靶向100多个位点。为了证实转染,构建并使用了c末端带有gfp的cas9蛋白。将h1299细胞在装有rpmi-1640(10%胎牛血清)的75t烧瓶中培养至约50%的融合度,胰蛋白酶消化,pbs洗涤,最后在于neon电穿孔缓冲液r中重悬。将150,000个细胞和由1.2μmcas9-gfp蛋白和1.5μm向导rna制成的rnp复合物加入10μlneon移液枪吸头中,在1,300v,20ms和2个脉冲的条件下进行电穿孔。然后,细胞在rpmi-1640(10%胎牛血清)培养基中恢复培养,两天后,根据使用普洛麦格的celltiterglo2.0测量的活细胞信号(荧光)确定活细胞数并相互比较。结果示于图30和32中。在图30至32中,与仅用蛋白质或引导rna引入的对照相比,在已知具有单一靶的ccr5靶实验组中未观察到明显的细胞凋亡。但是,在mt2、gnpda2和smim11实验组中观察到显著的细胞凋亡,这些实验组引入了导致裂解的rnp。具体地,用rnp引入的mt2、gnpda2和smim11实验组分别显示出约33%、约71%和约40%的凋亡率。通过以上实验,证实了当将cas9蛋白和细胞基因组序列特异性多靶标引导rna引入癌细胞中时,诱导了细胞凋亡并且可以通过rnp控制该作用。实施例20:确认使用cas12a蛋白的序列特异性凋亡效应和突变cnv序列特异性凋亡效应在该实施例中,已经证明使用cas12a蛋白表达载体可以在作为肺癌细胞的hcc827细胞的基因组中引起双重或多重切割,从而诱导凋亡。在本发明中,cas12a的dna和氨基酸序列由seqidnos:135和136表示。电穿孔用于将载体导入细胞和野生型非必需基因ccr5(seqidno:138)或crrna(seqidno:139)。使用靶向已知在hcc827细胞中存在18个以上拷贝的egfr_e2突变序列。将hcc827细胞在含有rpmi-1640(10%胎牛血清)培养基的75t烧瓶中培养至约50%的融合度,胰蛋白酶消化,pbs洗涤,最后在neon电转化缓冲液r中重悬。将150,000个细胞和500ng载体加载至10μlneon移液器吸头中,在1300v,20ms和2个脉冲的条件下进行电穿孔。然后,细胞在rpmi-1640(10%胎牛血清)培养基中恢复培养,6天后收获,并通过测量发光信号对细胞进行定量。另一方面,用作对照的egfr_wt序列(seqidno:137)是已知在hcc827细胞中不存在的序列。结果示于图33和34中。如图33和34所示,与对照组相比,在靶向ccr5的实验组中约76%的细胞被杀死,而在靶向egfr_e2的实验组中约83%的细胞被杀死。通过该实验证实,当使用cas12a蛋白切割癌细胞特异性序列而不考虑拷贝数时,可以诱导靶特异性凋亡。<110>g+flas生命科学有限公司<120>用于治疗癌症的包含向导rna和核酸内切酶作活性成分的药物组合物<130>pcb903027gfs<150>kr10-2018-0035298<151>2018-03-27<160>139<170>kopatentin2.0<210>1<211>19<212>dna<213>人工序列(artificialsequence)<400>1taatacgactcactatagg19<210>2<211>76<212>dna<213>人工序列(artificialsequence)<400>2gttttagagctagaaatagcaagttaaaataaggctagtccgttatcaacttgaaaaagt60ggcaccgagtcggtgc76<210>3<211>111<212>dna<213>人工序列(artificialsequence)<400>3taatacgactcactataggtgagaccgaggtctcggttttagagctagaaatagcaagtt60aaaataaggctagtccgttatcaacttgaaaaagtggcaccgagtcggtgc111<210>4<211>215<212>prt<213>人工序列(artificialsequence)<400>4metseraspasnalaglnleuthrglyleucysaspargphearggly151015phetyrprovalvalileaspvalgluthralaglypheasnalalys202530thraspalaleuleugluilealaalailethrleulysmetaspglu354045glnglytrpleumetproaspthrthrleuhisphehisvalglupro505560phevalglyalaasnleuglnproglualaleualapheasnglyile65707580aspproasnaspproaspargglyalavalserglutyrglualaleu859095hisgluilephelysvalvalarglysglyilelysalaserglycys100105110asnargalailemetvalalahisasnalaasnpheasphisserphe115120125metmetalaalaalagluargalaserleulysargasnprophehis130135140prophealathrpheaspthralaalaleualaglyleualaleugly145150155160glnthrvalleuserlysalacysglnthralaglymetasppheasp165170175serthrglnalahisseralaleutyraspthrgluargthralaval180185190leuphecysgluilevalasnargtrplysargleuglyglytrppro195200205leuseralaalaglugluval210215<210>5<211>236<212>prt<213>人工序列(artificialsequence)<400>5metserglualaproargalagluthrphevalpheleuaspleuglu151015alathrglyleuproservalgluprogluilealagluleuserleu202530phealavalhisargserserleugluasnprogluhisaspgluser354045glyalaleuvalleuproargvalleuasplysleuthrleucysmet505560cysprogluargprophethralalysalasergluilethrglyleu65707580sersergluglyleualaargcysarglysalaglypheaspglyala859095valvalargthrleuglnalapheleuserargglnalaglyproile100105110cysleuvalalahisasnglypheasptyrasppheproleuleucys115120125alagluleuargargleuglyalaargleuproargaspthrvalcys130135140leuaspthrleuproalaleuargglyleuaspargalahisserhis145150155160glythrargalaargglyargglnglytyrserleuglyserleuphe165170175hisargtyrpheargalagluproseralaalahisseralaglugly180185190aspvalhisthrleuleuleuilepheleuhisargalaalagluleu195200205leualatrpalaaspgluglnalaargglytrpalahisileglupro210215220mettyrleuproproaspaspproserleugluala225230235<210>6<211>304<212>prt<213>人工序列(artificialsequence)<400>6metglnthrleuilephepheaspmetglualathrglyleuprophe151015serglnprolysvalthrgluleucysleuleualavalhisargcys202530alaleugluserproprothrserglnglyproproprothrvalpro354045proproproargvalvalasplysleuserleucysvalalaprogly505560lysalacysserproalaalasergluilethrglyleuserthrala65707580valleualaalahisglyargglncyspheaspaspasnleualaasn859095leuleuleualapheleuargargglnproglnprotrpcysleuval100105110alahisasnglyaspargtyrasppheproleuleuglnalagluleu115120125alametleuglyleuthrseralaleuaspglyalaphecysvalasp130135140serilethralaleulysalaleugluargalaserserproserglu145150155160hisglyproarglyssertyrserleuglyseriletyrthrargleu165170175tyrglyglnserproproaspserhisthralagluglyaspvalleu180185190alaleuleuserilecysglntrpargproglnalaleuleuargtrp195200205valaspalahisalaargpropheglythrileargpromettyrgly210215220valthralaseralaargthrlysproargproseralavalthrthr225230235240thralahisleualathrthrargasnthrserproserleuargglu245250255serargglythrlysaspleuproprovallysaspproglyalaleu260265270serarggluglyleuleualaproleuglyleuleualaileleuthr275280285leualavalalathrleutyrglyleuserleualathrproglyasp290295300<210>7<211>1180<212>prt<213>人工序列(artificialsequence)<400>7metseraspvalalagluthrleuaspproleuargleuproleugln151015glygluargleuileglualaseralaglythrglylysthrphethr202530ilealaalaleutyrleuargleuleuleuglyleuglyglyserala354045alapheproargproleuthrvalglugluleuleuvalvalthrphe505560thrglualaalathralagluleuargglyargileargserasnile65707580hisgluleuargilealacysleuarggluthrthraspasnproleu859095tyrgluargleuleuglugluileaspasplysalaglnalaalagln100105110trpleuleuleualagluargglnmetaspglualaalavalphethr115120125ilehisglyphecysglnargmetleuasnleuasnalaphegluser130135140glymetleuphegluglnglnleuilegluaspgluserleuleuarg145150155160tyrglnalacysalaaspphetrpargarghiscystyrproleupro165170175arggluilealaglnvalvalphegluthrtrplysglyproglnala180185190leuleuargaspileasnargtyrleuglnglyglualaprovalile195200205lysalaproproproaspaspgluthrleualaserarghisalagln210215220ilevalalaargileaspthrvallysglnglntrpargaspalaval225230235240glygluleuaspalaleuilegluserserglyileaspargarglys245250255pheasnargserasnglnalalystrpileasplysileseralatrp260265270alagluglugluthrasnsertyrglnleuprogluserleuglulys275280285pheserglnargpheleugluaspargthrlysalaglyglygluthr290295300proarghisproleupheglualaileaspglnleuleualaglupro305310315320leuserileargaspleuvalilethrargalaleualagluilearg325330335gluthrvalalaargglulysargargargglygluleuglypheasp340345350aspmetleuserargleuaspseralaleuargsergluserglyglu355360365valleualaalaalaileargthrargpheprovalalametileasp370375380glupheglnaspthraspproglnglntyrargilepheargargile385390395400trphishisglnprogluthralaleuleuleuileglyaspprolys405410415glnalailetyralapheargglyalaaspilephethrtyrmetlys420425430alaargsergluvalhisalahistyrthrleuaspthrasntrparg435440445seralaproglymetvalasnservalasnlysleupheserglnthr450455460aspaspalaphemetphearggluilepropheileprovallysser465470475480alaglylysasnglnalaleuargphevalphelysglygluthrgln485490495proalametlysmettrpleumetgluglyglusercysglyvalgly500505510asptyrglnserthrmetalaglnvalcysalaalaglnileargasp515520525trpleuglnalaglyglnargglyglualaleuleumetasnglyasp530535540aspalaargprovalargalaseraspileservalleuvalargser545550555560argglnglualaalaglnvalargaspalaleuthrleuleugluile565570575proservaltyrleuserasnargaspservalphegluthrleuglu580585590alaglnglumetleutrpleuleuglnalavalmetthrprogluarg595600605gluasnthrleuargseralaleualathrsermetmetglyleuasn610615620alaleuaspilegluthrleuasnasnaspgluhisalatrpaspval625630635640valvalgluglupheaspglytyrargglniletrparglysarggly645650655valmetprometleuargalaleumetseralaargasnilealaglu660665670asnleuleualathralaglyglygluargargleuthraspileleu675680685hisilesergluleuleuglnglualaglythrglnleugluserglu690695700hisalaleuvalargtrpleuserglnhisileleugluproaspser705710715720asnalaserserglnglnmetargleugluserasplyshisleuval725730735glnilevalthrilehislysserlysglyleuglutyrproleuval740745750trpleupropheilethrasnpheargvalglngluglnalaphetyr755760765hisasparghisserpheglualavalleuaspleuasnalaalapro770775780gluservalaspleualaglualagluargleualagluaspleuarg785790795800leuleutyrvalalaleuthrargservaltrphiscysserleugly805810815valalaproleuvalargargargglyasplyslysglyaspthrasp820825830valhisglnseralaleuglyargleuleuglnlysglygluprogln835840845aspalaalaglyleuargthrcysileglualaleucysaspaspasp850855860ilealatrpglnthralaglnthrglyaspasnglnprotrpglnval865870875880asnaspvalserthralagluleuasnalalysthrleuglnargleu885890895proglyaspasntrpargvalthrsertyrserglyleuglnglnarg900905910glyhisglyilealaglnaspleumetproargleuaspvalaspala915920925alaglyvalalaservalvalglugluprothrleuthrprohisgln930935940pheproargglyalaserproglythrpheleuhisserleupheglu945950955960aspleuaspphethrglnprovalaspproasntrpvalargglulys965970975leugluleuglyglyphegluserglntrpgluprovalleuthrglu980985990trpilethralavalleuglnalaproleuasngluthrglyvalser99510001005leuserglnleuseralaargasnlysglnvalglumetgluphetyr101010151020leuproilesergluproleuilealaserglnleuaspthrleuile1025103010351040argglnpheaspproleuseralaglycysproproleugluphemet104510501055glnvalargglymetleulysglypheileaspleuvalphearghis106010651070gluglyargtyrtyrleuleuasptyrlysserasntrpleuglyglu107510801085aspserseralatyrthrglnglnalametalaalaalametglnala109010951100hisargtyraspleuglntyrglnleutyrthrleualaleuhisarg1105111011151120tyrleuarghisargilealaasptyrasptyrgluhishisphegly112511301135glyvaliletyrleupheleuargglyvalasplysgluhisprogln114011451150glnglyiletyrthrthrargproasnalaglyleuilealaleumet115511601165aspglumetphealaglymetthrleuglugluala117011751180<210>8<211>1122<212>prt<213>人工序列(artificialsequence)<400>8metleuargvaltyrhisserasnargleuaspvalleuglualaleu151015metglupheilevalgluarggluargleuaspasppropheglupro202530glumetileleuvalglnserthrglymetalaglntrpleuglnmet354045thrleuserglnlyspheglyilealaalaasnileasppheproleu505560proalaserpheiletrpaspmetphevalargvalleuprogluile65707580prolysgluseralapheasnlysglnsermetsertrplysleumet859095thrleuleuproglnleuleugluarggluaspphethrleuleuarg100105110histyrleuthraspaspserasplysarglysleupheglnleuser115120125serlysalaalaaspleupheaspglntyrleuvaltyrargproasp130135140trpleualaglntrpgluthrglyhisleuvalgluglyleuglyglu145150155160alaglnalatrpglnalaproleutrplysalaleuvalglutyrthr165170175hisglnleuglyglnproargtrphisargalaasnleutyrglnarg180185190pheilegluthrleugluseralathrthrcysproproglyleupro195200205serargvalpheilecysglyileseralaleuproprovaltyrleu210215220glnalaleuglnalaleuglylyshisilegluilehisleuleuphe225230235240thrasnprocysargtyrtyrtrpglyaspilelysaspproalatyr245250255leualalysleuleuthrargglnargarghisserphegluasparg260265270gluleuproleupheargaspsergluasnalaglyglnleupheasn275280285seraspglygluglnaspvalglyasnproleuleualasertrpgly290295300lysleuglyargasptyriletyrleuleuseraspleugluserser305310315320glngluleuaspalaphevalaspvalthrproaspasnleuleuhis325330335asnileglnseraspileleugluleugluasnargalavalalagly340345350valasnilegluglupheserargseraspasnlysargproleuasp355360365proleuaspserserilethrphehisvalcyshisserproglnarg370375380gluvalgluvalleuhisaspargleuleualametleuglugluasp385390395400prothrleuthrproargaspileilevalmetvalalaaspileasp405410415sertyrserpropheileglnalavalpheglyseralaproalaasp420425430argtyrleuprotyralaileseraspargargalaargglnserhis435440445provalleuglualapheileserleuleuserleuproaspserarg450455460phevalsergluaspvalleualaleuleuaspvalprovalleuala465470475480alaargpheaspilethrglugluglyleuargtyrleuargglntrp485490495valasngluserglyileargtrpglyileaspaspaspasnvalarg500505510gluleugluleuproalathrglyglnhisthrtrpargpheglyleu515520525thrargmetleuleuglytyralametgluseralaglnglyglutrp530535540glnservalleuprotyraspgluserserglyleuilealagluleu545550555560valglyhisleualaserleuleumetglnleuasniletrpargarg565570575glyleualaglngluargproleugluglutrpleuprovalcysarg580585590aspmetleuasnalaphepheleuproaspalagluthrglualaala595600605metthrleuilegluglnglntrpglnalaileilealagluglyleu610615620glyalaglntyrglyaspalavalproleuserleuleuargaspglu625630635640leualaglnargleuaspglngluargileserglnargpheleuala645650655glyprovalasnilecysthrleumetprometargserileprophe660665670lysvalvalcysleuleuglymetasnaspglyvaltyrproarggln675680685leualaproleuglypheaspleumetserglnlysprolysarggly690695700aspargserargargaspaspaspargtyrleupheleuglualaleu705710715720ileseralaglnglnlysleutyrilesertyrileglyargserile725730735glnaspasnsergluargpheproservalleuvalglngluleuile740745750asptyrileglyglnserhistyrleuproglyaspglualaleuasn755760765cysaspgluserglualaargvallysalahisleuthrcysleuhis770775780thrargmetpropheaspproglnasntyrglnproglygluarggln785790795800sertyralaargglutrpleuproalaalaserglnalaglylysala805810815hissergluphevalglnproleuprophethrleuprogluthrval820825830proleugluthrleuglnargphetrpalahisprovalargalaphe835840845pheglnmetargleuglnvalasnpheargthrgluaspsergluile850855860proaspthrglupropheileleugluglyleuserargtyrglnile865870875880asnglnglnleuleuasnalaleuvalgluglnaspaspalagluarg885890895leupheargargpheargalaalaglyaspleuprotyrglyalaphe900905910glygluilephetrpgluthrglncysglnglumetglnglnleuala915920925aspargvalilealacysargglnproglyglnsermetgluileasp930935940leualacysasnglyvalglnilethrglytrpleuproglnvalgln945950955960proaspglyleuleuargtrpargproserleuleuservalalagln965970975glymetglnleutrpleugluhisleuvaltyrcysalaserglygly980985990asnglygluserargleupheleuarglysaspglyglutrpargphe99510001005proproleualaalagluglnalaleuhistyrleuserglnleuile101010151020gluglytyrarggluglymetseralaproleuleuvalleuproglu1025103010351040serglyglyalatrpleulysthrcystyraspalaglnasnaspala104510501055metleuaspaspaspserthrleuglnlysalaargthrlyspheleu106010651070glnalatyrgluglyasnmetmetvalargglygluglyaspaspile107510801085trptyrglnargleutrpargglnleuthrprogluthrmetgluala109010951100ilevalgluglnserglnargpheleuleuproleupheargpheasn1105111011151120glnser<210>9<211>608<212>prt<213>人工序列(artificialsequence)<400>9metlysleuglnlysglnleuleuglualavalgluhislysglnleu151015argproleuaspvalglnphealaleuthrvalalaglyaspgluhis202530proalavalthrleualaalaalaleuleuserhisaspalaglyglu354045glyhisvalcysleuproleuserargleugluasnasnglualaser505560hisproleuleualathrcysvalsergluileglygluleuglnasn65707580trpgluglucysleuleualaserglnalavalserargglyaspglu859095prothrprometileleucysglyaspargleutyrleuasnargmet100105110trpcysasngluargthrvalalaargphepheasngluvalasnhis115120125alailegluvalaspglualaleuleualaglnthrleuasplysleu130135140pheprovalseraspgluileasntrpglnlysvalalaalaalaval145150155160alaleuthrargargileservalileserglyglyproglythrgly165170175lysthrthrthrvalalalysleuleualaalaleuileglnmetala180185190aspglygluargcysargileargleualaalaprothrglylysala195200205alaalaargleuthrgluserleuglylysalaleuargglnleupro210215220leuthraspgluglnlyslysargileprogluaspalaserthrleu225230235240hisargleuleuglyalaglnproglyserglnargleuarghishis245250255alaglyasnproleuhisleuaspvalleuvalvalaspglualaser260265270metileaspleuprometmetserargleuileaspalaleuproasp275280285hisalaargvalilepheleuglyaspargaspglnleualaserval290295300glualaglyalavalleuglyaspilecysalatyralaasnalagly305310315320phethralagluargalaargglnleuserargleuthrglythrhis325330335valproalaglythrglythrglualaalaserleuargaspserleu340345350cysleuleuglnlyssertyrargpheglyseraspserglyilegly355360365glnleualaalaalaileasnargglyasplysthralavallysthr370375380valpheglnglnaspphethraspileglulysargleuleuglnser385390395400glygluasptyrilealametleugluglualaleualaglytyrgly405410415argtyrleuaspleuleuglnalaargalagluproaspleuileile420425430glnalapheasnglutyrglnleuleucysalaleuarggluglypro435440445pheglyvalalaglyleuasngluargilegluglnphemetglngln450455460lysarglysilehisarghisprohisserargtrptyrgluglyarg465470475480provalmetilealaargasnaspseralaleuglyleupheasngly485490495aspileglyilealaleuaspargglyglnglythrargvaltrpphe500505510alametproaspglyasnilelysservalglnproserargleupro515520525gluhisgluthrthrtrpalametthrvalhislysserglnglyser530535540glupheasphisalaalaleuileleuproserglnargthrproval545550555560valthrarggluleuvaltyrthralavalthrargalaargargarg565570575leuserleutyralaaspgluargileleuseralaalailealathr580585590argthrgluargargserglyleualaalaleupheserserargglu595600605<210>10<211>475<212>prt<213>人工序列(artificialsequence)<400>10metmetasnaspglylysglnglnserthrpheleuphehisasptyr151015gluthrpheglythrhisproalaleuaspargproalaglnpheala202530alaileargthraspserglupheasnvalileglygluprogluval354045phetyrcyslysproalaaspasptyrleuproglnproglyalaval505560leuilethrglyilethrproglnglualaargalalysglygluasn65707580glualaalaphealaalaargilehisserleuphethrvalprolys859095thrcysileleuglytyrasnasnvalargpheaspaspgluvalthr100105110argasnilephetyrargasnphetyraspprotyralatrpsertrp115120125glnhisaspasnserargtrpaspleuleuaspvalmetargalacys130135140tyralaleuargprogluglyileasntrpprogluasnaspaspgly145150155160leuproserpheargleugluhisleuthrlysalaasnglyileglu165170175hisserasnalahisaspalametalaaspvaltyralathrileala180185190metalalysleuvallysthrargglnproargleupheasptyrleu195200205phethrhisargasnlyshislysleumetalaleuileaspvalpro210215220glnmetlysproleuvalhisvalserglymetpheglyalatrparg225230235240glyasnthrsertrpvalalaproleualatrphisprogluasnarg245250255asnalavalilemetvalaspleualaglyaspileserproleuleu260265270gluleuaspseraspthrleuarggluargleutyrthralalysthr275280285aspleuglyaspasnalaalavalprovallysleuvalhisileasn290295300lyscysprovalleualaglnalaasnthrleuargprogluaspala305310315320aspargleuglyileasnargglnhiscysleuaspasnleulysile325330335leuarggluasnproglnvalargglulysvalvalalailepheala340345350glualagluprophethrproseraspasnvalaspalaglnleutyr355360365asnglyphepheseraspalaaspargalaalametlysilevalleu370375380gluthrgluproargasnleuproalaleuaspilethrphevalasp385390395400lysargileglulysleuleupheasntyrargalaargasnphepro405410415glythrleuasptyralagluglnglnargtrpleugluhisargarg420425430glnvalphethrproglupheleuglnglytyralaaspgluleugln435440445metleuvalglnglntyralaaspasplysglulysvalalaleuleu450455460lysalaleutrpglntyralaglugluileval465470475<210>11<211>268<212>prt<213>人工序列(artificialsequence)<400>11metlysphevalserpheasnileasnglyleuargalaargprohis151015glnleuglualailevalglulyshisglnproaspvalileglyleu202530glngluthrlysvalhisaspaspmetpheproleuglugluvalala354045lysleuglytyrasnvalphetyrhisglyglnlysglyhistyrgly505560valalaleuleuthrlysgluthrproilealavalargargglyphe65707580proglyaspaspgluglualaglnargargileilemetalagluile859095proserleuleuglyasnvalthrvalileasnglytyrpheprogln100105110glygluserargasphisproilelyspheproalalysalaglnphe115120125tyrglnasnleuglnasntyrleugluthrgluleulysargaspasn130135140provalleuilemetglyaspmetasnileserprothraspleuasp145150155160ileglyileglyglugluasnarglysargtrpleuargthrglylys165170175cysserpheleuproglugluargglutrpmetaspargleumetser180185190trpglyleuvalaspthrphearghisalaasnproglnthralaasp195200205argphesertrppheasptyrargserlysglypheaspaspasnarg210215220glyleuargileaspleuleuleualaserglnproleualaglucys225230235240cysvalgluthrglyileasptyrgluileargsermetglulyspro245250255serasphisalaprovaltrpalathrpheargarg260265<210>12<211>355<212>prt<213>人工序列(artificialsequence)<400>12metglnthrleuglnmetserleuleuthrglnprotyrvalglnpro151015argpheprocyslysargtyrprothrpheseralasercysargthr202530glnlysthralailethrlysthrglulysvalphephesergluser354045pheaspglnthrargcysthrglnproleuserglulyslyslysarg505560valphepheleuaspvalasnproleucystyrgluglyserlyspro65707580serleuargserpheglyargtrpleuserleupheleuhisglnval859095serleuthraspprovalilealavalileaspglygluglyglyser100105110gluhisargarglysleuleuprosertyrlysalahisarglyslys115120125phemetarghismetserserglyhisvalglyargserhisglnval130135140ileasnaspvalleuglylyscysasnvalprovalilelysvalala145150155160glyhisglualaaspaspvalvalalathrleualaglyglnvalval165170175asnlysglypheargvalvalileglyserproasplysaspphelys180185190glnleuilesergluaspvalglnilevalmetproleuprogluleu195200205glnargtrpserphetyrthrleuarghistyrargaspglntyrasn210215220cysaspprogluseraspleuserpheargcysilevalglyaspglu225230235240valaspglyvalproglyileglnhisleuvalproserpheglyarg245250255lysthralametlysleuilelyslyshisglyserleugluthrleu260265270leuasnalaalaalaileargthrvalglyargprotyralaglnasp275280285alaleulysasnhisalaasptyrleuargargasntyrgluvalleu290295300alaleulysargaspvalasnileglnleutyraspglutrpleuval305310315320lysargaspasnhisasnasplysthralaleuserserphephelys325330335tyrleuglygluserlysgluleusertyrasnglyargproileser340345350tyrasngly355<210>13<211>577<212>prt<213>人工序列(artificialsequence)<400>13vallysglnglnileglnleuargargarggluvalaspgluthrala151015aspleuproalagluleuproproleuleuargargleutyralaser202530argglyvalargseralaglngluleugluargservallysglymet354045leuprotrpglnglnleuserglyvalglulysalavalgluileleu505560tyrasnalaphearggluglythrargileilevalvalglyaspphe65707580aspalaaspglyalathrserthralaleuservalleualametarg859095serleuglycysserasnileasptyrleuvalproasnargpheglu100105110aspglytyrglyleuserprogluvalvalaspglnalahisalaarg115120125glyalaglnleuilevalthrvalaspasnglyileserserhisala130135140glyvalgluhisalaargserleuglyileprovalilevalthrasp145150155160hishisleuproglyaspthrleuproalaalaglualaileileasn165170175proasnleuargaspcysasnpheproserlysserleualaglyval180185190glyvalalaphetyrleumetleualaleuargthrpheleuargasp195200205glnglytrppheaspgluargasnilealaileproasnleualaglu210215220leuleuaspleuvalalaleuglythrvalalaaspvalvalproleu225230235240aspalaasnasnargileleuthrtrpglnglymetserargilearg245250255alaglylyscysargproglyilelysalaleuleugluvalalaasn260265270argaspalaglnlysleualaalaseraspleuglyphealaleugly275280285proargleuasnalaalaglyargleuaspaspmetservalglyval290295300alaleuleuleucysaspasnileglyglualaargvalleualaasn305310315320gluleuaspalaleuasnglnthrarglysgluilegluglnglymet325330335glnileglualaleuthrleucysglulysleugluargserargasp340345350thrleuproglyglyleualamettyrhisproglutrphisglngly355360365valvalglyileleualaserargilelysgluargphehisargpro370375380valilealaphealaproalaglyaspglythrleulysglysergly385390395400argserileglnglyleuhismetargaspalaleugluargleuasp405410415thrleutyrproglymetmetleulyspheglyglyhisalametala420425430alaglyleuserleuglugluasplysphelysleupheglnglnarg435440445pheglygluleuvalthrglutrpleuaspproserleuleuglngly450455460gluvalvalseraspglyproleuserproalaglumetthrmetglu465470475480valalaglnleuleuargaspalaglyprotrpglyglnmetphepro485490495gluproleupheaspglyhispheargleuleuglnglnargleuval500505510glygluarghisleulysvalmetvalgluprovalglyglyglypro515520525leuleuaspglyilealapheasnvalaspthralaleutrpproasp530535540asnglyvalarggluvalglnleualatyrlysleuaspileasnglu545550555560pheargglyasnargserleuglnileileileaspasniletrppro565570575ile<210>14<211>866<212>prt<213>人工序列(artificialsequence)<400>14metserthrlysproleupheleuleuarglysalalyslysserser151015glygluproaspvalvalleutrpalaserasnaspphegluserthr202530cysalathrleuasptyrleuilevallysserglylyslysleuser354045sertyrphelysalavalalathrasnpheprovalvalasnaspleu505560proalagluglygluileaspphethrtrpsergluargtyrglnleu65707580serlysaspsermetthrtrpgluleulysproglyalaalaproasp859095asnalahistyrglnglyasnthrasnvalasnglygluaspmetthr100105110gluileglugluasnmetleuleuproileserglyglngluleupro115120125ileargtrpleualaglnhisglyserglulysprovalthrhisval130135140serargaspglyleuglnalaleuhisilealaargalaglugluleu145150155160proalavalthralaleualavalserhislysthrserleuleuasp165170175proleugluilearggluleuhislysleuvalargaspthrasplys180185190valpheproasnproglyasnserasnleuglyleuilethralaphe195200205pheglualatyrleuasnalaasptyrthraspargglyleuleuthr210215220lysglutrpmetlysglyasnargvalserhisilethrargthrala225230235240serglyalaasnalaglyglyglyasnleuthraspargglyglugly245250255phevalhisaspleuthrserleualaargaspvalalathrglyval260265270leualaargsermetaspleuaspiletyrasnleuhisproalahis275280285alalysargileglugluileilealagluasnlyspropropheser290295300valpheargasplyspheilethrmetproglyglyleuasptyrser305310315320argalailevalvalalaservallysglualaproileglyileglu325330335valileproalahisvalthrglutyrleuasnlysvalleuthrglu340345350thrasphisalaasnproaspprogluilevalaspilealacysgly355360365argserseralaprometproglnargvalthrglugluglylysgln370375380aspaspgluglulysproglnproserglythrthralavalglugln385390395400glyglualagluthrmetgluproaspalathrgluhishisglnasp405410415thrglnproleuaspalaglnserglnvalasnservalaspalalys420425430tyrglngluleuargalagluleuhisglualaarglysasnilepro435440445serlysasnprovalaspaspasplysleuleualaalaserarggly450455460gluphevalaspglyileseraspproasnaspprolystrpvallys465470475480glyileglnthrargaspcysvaltyrglnasnglnprogluthrglu485490495lysthrserproaspmetasnglnprogluprovalvalglnglnglu500505510progluilealacysasnalacysglyglnthrglyglyaspasncys515520525proaspcysglyalavalmetglyaspalathrtyrglngluthrphe530535540aspglugluserglnvalglualalysgluasnaspprogluglumet545550555560gluglyalagluhisprohisasngluasnalaglyseraspprohis565570575argaspcysseraspgluthrglygluvalalaaspprovalileval580585590gluaspilegluproglyiletyrtyrglyileserasngluasntyr595600605hisalaglyproglyileserlysserglnleuaspaspilealaasp610615620thrproalaleutyrleutrparglysasnalaprovalaspthrthr625630635640lysthrlysthrleuaspleuglythralaphehiscysargvalleu645650655gluprogluglupheserasnargpheilevalalaproglupheasn660665670argargthrasnalaglylysglugluglulysalapheleumetglu675680685cysalaserthrglylysthrvalilethralaglugluglyarglys690695700ilegluleumettyrglnservalmetalaleuproleuglyglntrp705710715720leuvalgluseralaglyhisalagluserseriletyrtrpgluasp725730735progluthrglyileleucysargcysargproasplysileilepro740745750gluphehistrpilemetaspvallysthrthralaaspileglnarg755760765phelysthralatyrtyrasptyrargtyrhisvalglnaspalaphe770775780tyrseraspglytyrglualaglnpheglyvalglnprothrpheval785790795800pheleuvalalaserthrthrileglucysglyargtyrprovalglu805810815ilephemetmetglygluglualalysleualaglyglnglnglutyr820825830hisargasnleuargthrleuseraspcysleuasnthraspglutrp835840845proalailelysthrleuserleuproargtrpalalysglutyrala850855860asnasp865<210>15<211>276<212>prt<213>人工序列(artificialsequence)<400>15metalaserargargasnleumetilevalaspglythrasnleugly151015pheargphelyshisasnasnserlyslysprophealasersertyr202530valserthrileglnserleualalyssertyrseralaargthrthr354045ilevalleuglyasplysglylysservalpheargleugluhisleu505560proglutyrlysglyasnargaspglulystyralaglnargthrglu65707580gluglulysalaleuaspgluglnphepheglutyrleulysaspala859095phegluleucyslysthrthrpheprothrphethrileargglyval100105110glualaaspaspmetalaalatyrilevallysleuileglyhisleu115120125tyrasphisvaltrpleuileserthraspglyasptrpaspthrleu130135140leuthrasplysvalserargpheserphethrthrargargglutyr145150155160hisleuargaspmettyrgluhishisasnvalaspaspvalglugln165170175pheileserleulysalailemetglyaspleuglyaspasnilearg180185190glyvalgluglyileglyalalysargglytyrasnileileargglu195200205pheglyasnvalleuaspileileaspglnleuproleuproglylys210215220glnlystyrileglnasnleuasnalaserglugluleuleuphearg225230235240asnleuileleuvalaspleuprothrtyrcysvalaspalaileala245250255alavalglyglnaspvalleuasplysphethrlysaspileleuglu260265270ilealaglugln275<210>16<211>226<212>prt<213>人工序列(artificialsequence)<400>16metthrproaspileileleuglnargthrglyileaspvalargala151015valgluglnglyaspaspalatrphislysleuargleuglyvalile202530thralasergluvalhisasnvalilealalysproargserglylys354045lystrpproaspmetlysmetsertyrphehisthrleuleualaglu505560valcysthrglyvalalaprogluvalasnalalysalaleualatrp65707580glylysglntyrgluasnaspalaargthrleuphegluphethrser859095glyvalasnvalthrgluserproileiletyrargaspglusermet100105110argthralacysserproaspglyleucysseraspglyasnglyleu115120125gluleulyscysprophethrserargaspphemetlyspheargleu130135140glyglypheglualailelysseralatyrmetalaglnvalglntyr145150155160sermettrpvalthrarglysasnalatrptyrphealaasntyrasp165170175proargmetlysarggluglyleuhistyrvalvalilegluargasp180185190glulystyrmetalaserpheaspgluilevalproglupheileglu195200205lysmetaspglualaleualagluileglyphevalpheglyglugln210215220trparg225<210>17<211>80<212>prt<213>人工序列(artificialsequence)<400>17metprolyslysasnglualaproalaserpheglulysalaleuser151015gluleugluglnilevalthrargleugluserglyaspleuproleu202530gluglualaleuasngluphegluargglyvalglnleualaarggln354045glyglnalalysleuglnglnalagluglnargvalglnileleuleu505560seraspasngluaspalaserleuthrprophethrproaspasnglu65707580<210>18<211>456<212>prt<213>人工序列(artificialsequence)<400>18metleuproserglnserproalailephethrvalserargleuasn151015glnthrvalargleuleuleugluhisglumetglyglnvaltrpile202530serglygluileserasnphethrglnproalaserglyhistrptyr354045phethrleulysaspaspthralaglnvalargcysalametphearg505560asnserasnargargvalthrpheargproglnhisglyglnglnval65707580leuvalargalaasnilethrleutyrgluproargglyasptyrgln859095ileilevalglusermetglnproalaglygluglyleuleuglngln100105110lystyrgluglnleulysalalysleuglnalagluglyleupheasp115120125glnglntyrlyslysproleuproserproalahiscysvalglyval130135140ilethrserlysthrglyalaalaleuhisaspileleuhisvalleu145150155160lysargargaspproserleuprovalileiletyrproalaalaval165170175glnglyaspaspalaproglyglnilevalargalailegluleuala180185190asnglnargasnglucysaspvalleuilevalglyargglyglygly195200205serleugluaspleutrpserpheasnaspgluargvalalaargala210215220ilephethrserargileprovalvalseralavalglyhisgluthr225230235240aspvalthrilealaaspphevalalaaspleuargalaprothrpro245250255seralaalaalagluvalvalserargasnglnglngluleuleuarg260265270glnvalglnserthrargglnargleuglumetalametasptyrtyr275280285leualaasnargthrargargphethrglnilehishisargleugln290295300glnglnhisproglnleuargleualaargglnglnthrmetleuglu305310315320argleuglnlysargmetserphealaleugluasnglnleulysarg325330335thrglyglnglnglnglnargleuthrglnargleuasnglnglnasn340345350proglnprolysilehisargalaglnthrargileglnglnleuglu355360365tyrargleualagluthrleuargalaglnleuseralathrargglu370375380argpheglyasnalavalthrhisleuglualavalserproleuser385390395400thrleualaargglytyrservalthrthralathraspglyasnval405410415leulyslysvallysglnvallysalaglyglumetleuthrthrarg420425430leugluaspglytrpileglusergluvallysasnileglnproval435440445lyslysserarglyslysvalhis450455<210>19<211>4101<212>dna<213>人工序列(artificialsequence)<400>19gacaagaagtacagcatcggcctggacatcggcaccaactctgtgggctgggccgtgatc60accgacgagtacaaggtgcccagcaagaaattcaaggtgctgggcaacaccgaccggcac120agcatcaagaagaacctgatcggagccctgctgttcgacagcggcgaaacagccgaggcc180acccggctgaagagaaccgccagaagaagatacaccagacggaagaaccggatctgctat240ctgcaagagatcttcagcaacgagatggccaaggtggacgacagcttcttccacagactg300gaagagtccttcctggtggaagaggataagaagcacgagcggcaccccatcttcggcaac360atcgtggacgaggtggcctaccacgagaagtaccccaccatctaccacctgagaaagaaa420ctggtggacagcaccgacaaggccgacctgcggctgatctatctggccctggcccacatg480atcaagttccggggccacttcctgatcgagggcgacctgaaccccgacaacagcgacgtg540gacaagctgttcatccagctggtgcagacctacaaccagctgttcgaggaaaaccccatc600aacgccagcggcgtggacgccaaggccatcctgtctgccagactgagcaagagcagacgg660ctggaaaatctgatcgcccagctgcccggcgagaagaagaatggcctgttcggaaacctg720attgccctgagcctgggcctgacccccaacttcaagagcaacttcgacctggccgaggat780gccaaactgcagctgagcaaggacacctacgacgacgacctggacaacctgctggcccag840atcggcgaccagtacgccgacctgtttctggccgccaagaacctgtccgacgccatcctg900ctgagcgacatcctgagagtgaacaccgagatcaccaaggcccccctgagcgcctctatg960atcaagagatacgacgagcaccaccaggacctgaccctgctgaaagctctcgtgcggcag1020cagctgcctgagaagtacaaagagattttcttcgaccagagcaagaacggctacgccggc1080tacattgacggcggagccagccaggaagagttctacaagttcatcaagcccatcctggaa1140aagatggacggcaccgaggaactgctcgtgaagctgaacagagaggacctgctgcggaag1200cagcggaccttcgacaacggcagcatcccccaccagatccacctgggagagctgcacgcc1260attctgcggcggcaggaagatttttacccattcctgaaggacaaccgggaaaagatcgag1320aagatcctgaccttccgcatcccctactacgtgggccctctggccaggggaaacagcaga1380ttcgcctggatgaccagaaagagcgaggaaaccatcaccccctggaacttcgaggaagtg1440gtggacaagggcgcttccgcccagagcttcatcgagcggatgaccaacttcgataagaac1500ctgcccaacgagaaggtgctgcccaagcacagcctgctgtacgagtacttcaccgtgtat1560aacgagctgaccaaagtgaaatacgtgaccgagggaatgagaaagcccgccttcctgagc1620ggcgagcagaaaaaggccatcgtggacctgctgttcaagaccaaccggaaagtgaccgtg1680aagcagctgaaagaggactacttcaagaaaatcgagtgcttcgactccgtggaaatctcc1740ggcgtggaagatcggttcaacgcctccctgggcacataccacgatctgctgaaaattatc1800aaggacaaggacttcctggacaatgaggaaaacgaggacattctggaagatatcgtgctg1860accctgacactgtttgaggacagagagatgatcgaggaacggctgaaaacctatgcccac1920ctgttcgacgacaaagtgatgaagcagctgaagcggcggagatacaccggctggggcagg1980ctgagccggaagctgatcaacggcatccgggacaagcagtccggcaagacaatcctggat2040ttcctgaagtccgacggcttcgccaacagaaacttcatgcagctgatccacgacgacagc2100ctgacctttaaagaggacatccagaaagcccaggtgtccggccagggcgatagcctgcac2160gagcacattgccaatctggccggcagccccgccattaagaagggcatcctgcagacagtg2220aaggtggtggacgagctcgtgaaagtgatgggccggcacaagcccgagaacatcgtgatc2280gaaatggccagagagaaccagaccacccagaagggacagaagaacagccgcgagagaatg2340aagcggatcgaagagggcatcaaagagctgggcagccagatcctgaaagaacaccccgtg2400gaaaacacccagctgcagaacgagaagctgtacctgtactacctgcagaatgggcgggat2460atgtacgtggaccaggaactggacatcaaccggctgtccgactacgatgtggaccatatc2520gtgcctcagagctttctgaaggacgactccatcgacaacaaggtgctgaccagaagcgac2580aagaaccggggcaagagcgacaacgtgccctccgaagaggtcgtgaagaagatgaagaac2640tactggcggcagctgctgaacgccaagctgattacccagagaaagttcgacaatctgacc2700aaggccgagagaggcggcctgagcgaactggataaggccggcttcatcaagagacagctg2760gtggaaacccggcagatcacaaagcacgtggcacagatcctggactcccggatgaacact2820aagtacgacgagaatgacaagctgatccgggaagtgaaagtgatcaccctgaagtccaag2880ctggtgtccgatttccggaaggatttccagttttacaaagtgcgcgagatcaacaactac2940caccacgcccacgacgcctacctgaacgccgtcgtgggaaccgccctgatcaaaaagtac3000cctaagctggaaagcgagttcgtgtacggcgactacaaggtgtacgacgtgcggaagatg3060atcgccaagagcgagcaggaaatcggcaaggctaccgccaagtacttcttctacagcaac3120atcatgaactttttcaagaccgagattaccctggccaacggcgagatccggaagcggcct3180ctgatcgagacaaacggcgaaaccggggagatcgtgtgggataagggccgggattttgcc3240accgtgcggaaagtgctgagcatgccccaagtgaatatcgtgaaaaagaccgaggtgcag3300acaggcggcttcagcaaagagtctatcctgcccaagaggaacagcgataagctgatcgcc3360agaaagaaggactgggaccctaagaagtacggcggcttcgacagccccaccgtggcctat3420tctgtgctggtggtggccaaagtggaaaagggcaagtccaagaaactgaagagtgtgaaa3480gagctgctggggatcaccatcatggaaagaagcagcttcgagaagaatcccatcgacttt3540ctggaagccaagggctacaaagaagtgaaaaaggacctgatcatcaagctgcctaagtac3600tccctgttcgagctggaaaacggccggaagagaatgctggcctctgccggcgaactgcag3660aagggaaacgaactggccctgccctccaaatatgtgaacttcctgtacctggccagccac3720tatgagaagctgaagggctcccccgaggataatgagcagaaacagctgtttgtggaacag3780cacaagcactacctggacgagatcatcgagcagatcagcgagttctccaagagagtgatc3840ctggccgacgctaatctggacaaagtgctgtccgcctacaacaagcaccgggataagccc3900atcagagagcaggccgagaatatcatccacctgtttaccctgaccaatctgggagcccct3960gccgccttcaagtactttgacaccaccatcgaccggaagaggtacaccagcaccaaagag4020gtgctggacgccaccctgatccaccagagcatcaccggcctgtacgagacacggatcgac4080ctgtctcagctgggaggcgac4101<210>20<211>1450<212>prt<213>人工序列(artificialsequence)<400>20asptyrlysasphisaspglyasptyrlysasphisaspileasptyr151015lysaspaspaspasplysmetgluthralaprolyslyslysarglys202530valglyilehisglyvalproalaalaasplyslystyrserilegly354045leuaspileglythrasnservalglytrpalavalilethraspglu505560tyrlysvalproserlyslysphelysvalleuglyasnthrasparg65707580hisserilelyslysasnleuileglyalaleuleupheaspsergly859095gluthralaglualathrargleulysargthralaargargargtyr100105110thrargarglysasnargilecystyrleuglngluilepheserasn115120125glumetgluthralalysvalaspaspserphephehisargleuglu130135140gluserpheleuvalglugluasplyslyshisgluarghisproile145150155160pheglyasnilevalaspgluvalalatyrhisglulystyrprothr165170175iletyrhisleuarglyslysleuvalaspserthrasplysalaasp180185190leuargleuiletyrleualaleualahismetgluthrilelysphe195200205argglyhispheleuilegluglyaspleuasnproaspasnserasp210215220valasplysleupheileglnleuvalglnthrtyrasnglnleuphe225230235240glugluasnproileasnalaserglyvalaspalalysalaileleu245250255seralaargleuserlysserargargleugluasnleuilealagln260265270leuproglyglulyslysasnglyleupheglyasnleuilealaleu275280285serleuglyleuthrproasnphelysserasnpheaspleualaglu290295300aspalalysleuglnleuserlysaspthrtyraspaspaspleuasp305310315320asnleuleualaglnileglyaspglntyralaaspleupheleuala325330335alalysasnleuseraspalaileleuleuseraspileleuargval340345350asnthrgluilethrlysalaproleuseralasermetgluthrile355360365lysargtyraspgluhishisglnaspleuthrleuleulysalaleu370375380valargglnglnleuproglulystyrlysgluilephepheaspgln385390395400serlysasnglytyralaglytyrileaspglyglyalaserglnglu405410415gluphetyrlyspheilelysproileleuglulysmetgluthrasp420425430glythrglugluleuleuvallysleuasnarggluaspleuleuarg435440445lysglnargthrpheaspasnglyserileprohisglnilehisleu450455460glygluleuhisalaileleuargargglngluaspphetyrprophe465470475480leulysaspasnargglulysileglulysileleuthrpheargile485490495protyrtyrvalglyproleualaargglyasnserargphealatrp500505510metgluthrthrarglysserglugluthrilethrprotrpasnphe515520525glugluvalvalasplysglyalaseralaglnserpheilegluarg530535540metgluthrthrasnpheasplysasnleuproasnglulysvalleu545550555560prolyshisserleuleutyrglutyrphethrvaltyrasngluleu565570575thrlysvallystyrvalthrgluglymetgluthrarglysproala580585590pheleuserglygluglnlyslysalailevalaspleuleuphelys595600605thrasnarglysvalthrvallysglnleulysgluasptyrphelys610615620lysileglucyspheaspservalgluileserglyvalgluasparg625630635640pheasnalaserleuglythrtyrhisaspleuleulysileilelys645650655asplysasppheleuaspasnglugluasngluaspileleugluasp660665670ilevalleuthrleuthrleuphegluaspargglumetgluthrile675680685glugluargleulysthrtyralahisleupheaspasplysvalmet690695700gluthrlysglnleulysargargargtyrthrglytrpglyargleu705710715720serarglysleuileasnglyileargasplysglnserglylysthr725730735ileleuasppheleulysseraspglyphealaasnargasnphemet740745750gluthrglnleuilehisaspaspserleuthrphelysgluaspile755760765glnlysalaglnvalserglyglnglyaspserleuhisgluhisile770775780alaasnleualaglyserproalailelyslysglyileleuglnthr785790795800vallysvalvalaspgluleuvallysvalmetgluthrglyarghis805810815lysprogluasnilevalileglumetgluthralaarggluasngln820825830thrthrglnlysglyglnlysasnserarggluargmetgluthrlys835840845argileglugluglyilelysgluleuglyserglnileleulysglu850855860hisprovalgluasnthrglnleuglnasnglulysleutyrleutyr865870875880tyrleuglnasnglyargaspmetgluthrtyrvalaspglngluleu885890895aspileasnargleuserasptyraspvalasphisilevalprogln900905910serpheleulysaspaspserileaspasnlysvalleuthrargser915920925asplysasnargglylysseraspasnvalproserglugluvalval930935940lyslysmetgluthrlysasntyrtrpargglnleuleuasnalalys945950955960leuilethrglnarglyspheaspasnleuthrlysalagluarggly965970975glyleusergluleuasplysalaglypheilelysargglnleuval980985990gluthrargglnilethrlyshisvalalaglnileleuaspserarg99510001005metgluthrasnthrlystyraspgluasnasplysleuileargglu101010151020vallysvalilethrleulysserlysleuvalseraspphearglys1025103010351040asppheglnphetyrlysvalarggluileasnasntyrhishisala104510501055hisaspalatyrleuasnalavalvalglythralaleuilelyslys106010651070tyrprolysleuglusergluphevaltyrglyasptyrlysvaltyr107510801085aspvalarglysmetgluthrilealalyssergluglngluilegly109010951100lysalathralalystyrphephetyrserasnilemetgluthrasn1105111011151120phephelysthrgluilethrleualaasnglygluilearglysarg112511301135proleuilegluthrasnglygluthrglygluilevaltrpasplys114011451150glyargaspphealathrvalarglysvalleusermetgluthrpro115511601165glnvalasnilevallyslysthrgluvalglnthrglyglypheser117011751180lysgluserileleuprolysargasnserasplysleuilealaarg1185119011951200lyslysasptrpaspprolyslystyrglyglypheaspserprothr120512101215valalatyrservalleuvalvalalalysvalglulysglylysser122012251230lyslysleulysservallysgluleuleuglyilethrilemetglu123512401245thrgluargserserpheglulysasnproileasppheleugluala125012551260lysglytyrlysgluvallyslysaspleuileilelysleuprolys1265127012751280tyrserleuphegluleugluasnglyarglysargmetgluthrleu128512901295alaseralaglygluleuglnlysglyasngluleualaleuproser130013051310lystyrvalasnpheleutyrleualaserhistyrglulysleulys131513201325glyserprogluaspasngluglnlysglnleuphevalgluglnhis133013351340lyshistyrleuaspgluileilegluglnileserglupheserlys1345135013551360argvalileleualaaspalaasnleuasplysvalleuseralatyr136513701375asnlyshisargasplysproilearggluglnalagluasnileile138013851390hisleuphethrleuthrasnleuglyalaproalaalaphelystyr139514001405pheaspthrthrileasparglysargtyrthrserthrlysgluval141014151420leuaspalathrleuilehisglnserilethrglyleutyrgluthr1425143014351440argileaspleuserglnleuglyglyasp14451450<210>21<211>3903<212>dna<213>人工序列(artificialsequence)<400>21atgtcaatttatcaagaatttgttaataaatatagtttaagtaaaactctaagatttgag60ttaatcccacagggtaaaacacttgaaaacataaaagcaagaggtttgattttagatgat120gagaaaagagctaaagactacaaaaaggctaaacaaataattgataaatatcatcagttt180tttatagaggagatattaagttcggtttgtattagcgaagatttattacaaaactattct240gatgtttattttaaacttaaaaagagtgatgatgataatctacaaaaagattttaaaagt300gcaaaagatacgataaagaaacaaatatctgaatatataaaggactcagagaaatttaag360aatttgtttaatcaaaaccttatcgatgctaaaaaagggcaagagtcagatttaattcta420tggctaaagcaatctaaggataatggtatagaactatttaaagccaatagtgatatcaca480gatatagatgaggcgttagaaataatcaaatcttttaaaggttggacaacttattttaag540ggttttcatgaaaatagaaaaaatgtttatagtagcaatgatattcctacatctattatt600tataggatagtagatgataatttgcctaaatttctagaaaataaagctaagtatgagagt660ttaaaagacaaagctccagaagctataaactatgaacaaattaaaaaagatttggcagaa720gagctaacctttgatattgactacaaaacatctgaagttaatcaaagagttttttcactt780gatgaagtttttgagatagcaaactttaataattatctaaatcaaagtggtattactaaa840tttaatactattattggtggtaaatttgtaaatggtgaaaatacaaagagaaaaggtata900aatgaatatataaatctatactcacagcaaataaatgataaaacactcaaaaaatataaa960atgagtgttttatttaagcaaattttaagtgatacagaatctaaatcttttgtaattgat1020aagttagaagatgatagtgatgtagttacaacgatgcaaagtttttatgagcaaatagca1080gcttttaaaacagtagaagaaaaatctattaaagaaacactatctttattatttgatgat1140ttaaaagctcaaaaacttgatttgagtaaaatttattttaaaaatgataaatctcttact1200gatctatcacaacaagtttttgatgattatagtgttattggtacagcggtactagaatat1260ataactcaacaaatagcacctaaaaatcttgataaccctagtaagaaagagcaagaatta1320atagccaaaaaaactgaaaaagcaaaatacttatctctagaaactataaagcttgcctta1380gaagaatttaataagcatagagatatagataaacagtgtaggtttgaagaaatacttgca1440aactttgcggctattccgatgatatttgatgaaatagctcaaaacaaagacaatttggca1500cagatatctatcaaatatcaaaatcaaggtaaaaaagacctacttcaagctagtgcggaa1560gatgatgttaaagctatcaaggatcttttagatcaaactaataatctcttacataaacta1620aaaatatttcatattagtcagtcagaagataaggcaaatattttagacaaggatgagcat1680ttttatctagtatttgaggagtgctactttgagctagcgaatatagtgcctctttataac1740aaaattagaaactatataactcaaaagccatatagtgatgagaaatttaagctcaatttt1800gagaactcgactttggctaatggttgggataaaaataaagagcctgacaatacggcaatt1860ttatttatcaaagatgataaatattatctgggtgtgatgaataagaaaaataacaaaata1920tttgatgataaagctatcaaagaaaataaaggcgagggttataaaaaaattgtttataaa1980cttttacctggcgcaaataaaatgttacctaaggttttcttttctgctaaatctataaaa2040ttttataatcctagtgaagatatacttagaataagaaatcattccacacatacaaaaaat2100ggtagtcctcaaaaaggatatgaaaaatttgagtttaatattgaagattgccgaaaattt2160atagatttttataaacagtctataagtaagcatccggagtggaaagattttggatttaga2220ttttctgatactcaaagatataattctatagatgaattttatagagaagttgaaaatcaa2280ggctacaaactaacttttgaaaatatatcagagagctatattgatagcgtagttaatcag2340ggtaaattgtacctattccaaatctataataaagatttttcagcttatagcaaagggcga2400ccaaatctacatactttatattggaaagcgctgtttgatgagagaaatcttcaagatgtg2460gtttataagctaaatggtgaggcagagcttttttatcgtaaacaatcaatacctaaaaaa2520atcactcacccagctaaagaggcaatagctaataaaaacaaagataatcctaaaaaagag2580agtgtttttgaatatgatttaatcaaagataaacgctttactgaagataagtttttcttt2640cactgtcctattacaatcaattttaaatctagtggagctaataagtttaatgatgaaatc2700aatttattgctaaaagaaaaagcaaatgatgttcatatattaagtatagatagaggtgaa2760agacatttagcttactatactttggtagatggtaaaggcaatatcatcaaacaagatact2820ttcaacatcattggtaatgatagaatgaaaacaaactaccatgataagcttgctgcaata2880gagaaagatagggattcagctaggaaagactggaaaaagataaataacatcaaagagatg2940aaagagggctatctatctcaggtagttcatgaaatagctaagctagttatagagtataat3000gctattgtggtttttgaggatttaaattttggatttaaaagagggcgtttcaaggtagag3060aagcaggtctatcaaaagttagaaaaaatgctaattgagaaactaaactatctagttttc3120aaagataatgagtttgataaaactgggggagtgcttagagcttatcagctaacagcacct3180tttgagacttttaaaaagatgggtaaacaaacaggtattatctactatgtaccagctggt3240tttacttcaaaaatttgtcctgtaactggttttgtaaatcagttatatcctaagtatgaa3300agtgtcagcaaatctcaagagttctttagtaagtttgacaagatttgttataaccttgat3360aagggctattttgagtttagttttgattataaaaactttggtgacaaggctgccaaaggc3420aagtggactatagctagctttgggagtagattgattaactttagaaattcagataaaaat3480cataattgggatactcgagaagtttatccaactaaagagttggagaaattgctaaaagat3540tattctatcgaatatgggcatggcgaatgtatcaaagcagctatttgcggtgagagcgac3600aaaaagttttttgctaagctaactagtgtcctaaatactatcttacaaatgcgtaactca3660aaaacaggtactgagttagattatctaatttcaccagtagcagatgtaaatggcaatttc3720tttgattcgcgacaggcgccaaaaaatatgcctcaagatgctgatgccaatggtgcttat3780catattgggctaaaaggtctgatgctactaggtaggatcaaaaataatcaagagggcaaa3840aaactcaatttggttatcaaaaatgaagagtattttgagttcgtgcagaataggaataac3900taa3903<210>22<211>1300<212>prt<213>人工序列(artificialsequence)<400>22metseriletyrglngluphevalasnlystyrserleuserlysthr151015leuargphegluleuileproglnglylysthrleugluasnilelys202530alaargglyleuileleuaspaspglulysargalalysasptyrlys354045lysalalysglnileileasplystyrhisglnphepheilegluglu505560ileleuserservalcysilesergluaspleuleuglnasntyrser65707580aspvaltyrphelysleulyslysseraspaspaspasnleuglnlys859095aspphelysseralalysaspthrilelyslysglnileserglutyr100105110ilelysaspserglulysphelysasnleupheasnglnasnleuile115120125aspalalyslysglyglngluseraspleuileleutrpleulysgln130135140serlysaspasnglyilegluleuphelysalaasnseraspilethr145150155160aspileaspglualaleugluileilelysserphelysglytrpthr165170175thrtyrphelysglyphehisgluasnarglysasnvaltyrserser180185190asnaspileprothrserileiletyrargilevalaspaspasnleu195200205prolyspheleugluasnlysalalystyrgluserleulysasplys210215220alaproglualaileasntyrgluglnilelyslysaspleualaglu225230235240gluleuthrpheaspileasptyrlysthrsergluvalasnglnarg245250255valpheserleuaspgluvalphegluilealaasnpheasnasntyr260265270leuasnglnserglyilethrlyspheasnthrileileglyglylys275280285phevalasnglygluasnthrlysarglysglyileasnglutyrile290295300asnleutyrserglnglnileasnasplysthrleulyslystyrlys305310315320metservalleuphelysglnileleuseraspthrgluserlysser325330335phevalileasplysleugluaspaspseraspvalvalthrthrmet340345350glnserphetyrgluglnilealaalaphelysthrvalgluglulys355360365serilelysgluthrleuserleuleupheaspaspleulysalagln370375380lysleuaspleuserlysiletyrphelysasnasplysserleuthr385390395400aspleuserglnglnvalpheaspasptyrservalileglythrala405410415valleuglutyrilethrglnglnilealaprolysasnleuaspasn420425430proserlyslysgluglngluleuilealalyslysthrglulysala435440445lystyrleuserleugluthrilelysleualaleugluglupheasn450455460lyshisargaspileasplysglncysargpheglugluileleuala465470475480asnphealaalaileprometilepheaspgluilealaglnasnlys485490495aspasnleualaglnileserilelystyrglnasnglnglylyslys500505510aspleuleuglnalaseralagluaspaspvallysalailelysasp515520525leuleuaspglnthrasnasnleuleuhislysleulysilephehis530535540ileserglnsergluasplysalaasnileleuasplysaspgluhis545550555560phetyrleuvalphegluglucystyrphegluleualaasnileval565570575proleutyrasnlysileargasntyrilethrglnlysprotyrser580585590aspglulysphelysleuasnphegluasnserthrleualaasngly595600605trpasplysasnlysgluproaspasnthralaileleupheilelys610615620aspasplystyrtyrleuglyvalmetasnlyslysasnasnlysile625630635640pheaspasplysalailelysgluasnlysglygluglytyrlyslys645650655ilevaltyrlysleuleuproglyalaasnlysmetleuprolysval660665670phepheseralalysserilelysphetyrasnprosergluaspile675680685leuargileargasnhisserthrhisthrlysasnglyserprogln690695700lysglytyrglulyspheglupheasnilegluaspcysarglysphe705710715720ileaspphetyrlysglnserileserlyshisproglutrplysasp725730735pheglypheargpheseraspthrglnargtyrasnserileaspglu740745750phetyrarggluvalgluasnglnglytyrlysleuthrphegluasn755760765ileserglusertyrileaspservalvalasnglnglylysleutyr770775780leupheglniletyrasnlysasppheseralatyrserlysglyarg785790795800proasnleuhisthrleutyrtrplysalaleupheaspgluargasn805810815leuglnaspvalvaltyrlysleuasnglyglualagluleuphetyr820825830arglysglnserileprolyslysilethrhisproalalysgluala835840845ilealaasnlysasnlysaspasnprolyslysgluservalpheglu850855860tyraspleuilelysasplysargphethrgluasplysphephephe865870875880hiscysproilethrileasnphelysserserglyalaasnlysphe885890895asnaspgluileasnleuleuleulysglulysalaasnaspvalhis900905910ileleuserileaspargglygluarghisleualatyrtyrthrleu915920925valaspglylysglyasnileilelysglnaspthrpheasnileile930935940glyasnaspargmetlysthrasntyrhisasplysleualaalaile945950955960glulysaspargaspseralaarglysasptrplyslysileasnasn965970975ilelysglumetlysgluglytyrleuserglnvalvalhisgluile980985990alalysleuvalileglutyrasnalailevalvalphegluaspleu99510001005asnpheglyphelysargglyargphelysvalglulysglnvaltyr101010151020glnlysleuglulysmetleuileglulysleuasntyrleuvalphe1025103010351040lysaspasnglupheasplysthrglyglyvalleuargalatyrgln104510501055leuthralaprophegluthrphelyslysmetglylysglnthrgly106010651070ileiletyrtyrvalproalaglyphethrserlysilecysproval107510801085thrglyphevalasnglnleutyrprolystyrgluservalserlys109010951100serglngluphepheserlyspheasplysilecystyrasnleuasp1105111011151120lysglytyrpheglupheserpheasptyrlysasnpheglyasplys112511301135alaalalysglylystrpthrilealaserpheglyserargleuile114011451150asnpheargasnserasplysasnhisasntrpaspthrarggluval115511601165tyrprothrlysgluleuglulysleuleulysasptyrserileglu117011751180tyrglyhisglyglucysilelysalaalailecysglygluserasp1185119011951200lyslysphephealalysleuthrservalleuasnthrileleugln120512101215metargasnserlysthrglythrgluleuasptyrleuileserpro122012251230valalaaspvalasnglyasnphepheaspserargglnalaprolys123512401245asnmetproglnaspalaaspalaasnglyalatyrhisileglyleu125012551260lysglyleumetleuleuglyargilelysasnasnglngluglylys1265127012751280lysleuasnleuvalilelysasngluglutyrphegluphevalgln128512901295asnargasnasn1300<210>23<211>3459<212>dna<213>人工序列(artificialsequence)<400>23atgaaagtgaccaaggtcgacggcatcagccacaagaagtacatcgaagagggcaagctc60gtgaagtccaccagcgaggaaaaccggaccagcgagagactgagcgagctgctgagcatc120cggctggacatctacatcaagaaccccgacaacgcctccgaggaagagaaccggatcaga180agagagaacctgaagaagttctttagcaacaaggtgctgcacctgaaggacagcgtgctg240tatctgaagaaccggaaagaaaagaacgccgtgcaggacaagaactatagcgaagaggac300atcagcgagtacgacctgaaaaacaagaacagcttctccgtgctgaagaagatcctgctg360aacgaggacgtgaactctgaggaactggaaatctttcggaaggacgtggaagccaagctg420aacaagatcaacagcctgaagtacagcttcgaagagaacaaggccaactaccagaagatc480aacgagaacaacgtggaaaaagtgggcggcaagagcaagcggaacatcatctacgactac540tacagagagagcgccaagcgcaacgactacatcaacaacgtgcaggaagccttcgacaag600ctgtataagaaagaggatatcgagaaactgtttttcctgatcgagaacagcaagaagcac660gagaagtacaagatccgcgagtactatcacaagatcatcggccggaagaacgacaaagag720aacttcgccaagattatctacgaagagatccagaacgtgaacaacatcaaagagctgatt780gagaagatccccgacatgtctgagctgaagaaaagccaggtgttctacaagtactacctg840gacaaagaggaactgaacgacaagaatattaagtacgccttctgccacttcgtggaaatc900gagatgtcccagctgctgaaaaactacgtgtacaagcggctgagcaacatcagcaacgat960aagatcaagcggatcttcgagtaccagaatctgaaaaagctgatcgaaaacaaactgctg1020aacaagctggacacctacgtgcggaactgcggcaagtacaactactatctgcaagtgggc1080gagatcgccacctccgactttatcgcccggaaccggcagaacgaggccttcctgagaaac1140atcatcggcgtgtccagcgtggcctacttcagcctgaggaacatcctggaaaccgagaac1200gagaacgatatcaccggccggatgcggggcaagaccgtgaagaacaacaagggcgaagag1260aaatacgtgtccggcgaggtggacaagatctacaatgagaacaagcagaacgaagtgaaa1320gaaaatctgaagatgttctacagctacgacttcaacatggacaacaagaacgagatcgag1380gacttcttcgccaacatcgacgaggccatcagcagcatcagacacggcatcgtgcacttc1440aacctggaactggaaggcaaggacatcttcgccttcaagaatatcgcccccagcgagatc1500tccaagaagatgtttcagaacgaaatcaacgaaaagaagctgaagctgaaaatcttcaag1560cagctgaacagcgccaacgtgttcaactactacgagaaggatgtgatcatcaagtacctg1620aagaataccaagttcaacttcgtgaacaaaaacatccccttcgtgcccagcttcaccaag1680ctgtacaacaagattgaggacctgcggaataccctgaagtttttttggagcgtgcccaag1740gacaaagaagagaaggacgcccagatctacctgctgaagaatatctactacggcgagttc1800ctgaacaagttcgtgaaaaactccaaggtgttctttaagatcaccaatgaagtgatcaag1860attaacaagcagcggaaccagaaaaccggccactacaagtatcagaagttcgagaacatc1920gagaaaaccgtgcccgtggaatacctggccatcatccagagcagagagatgatcaacaac1980caggacaaagaggaaaagaatacctacatcgactttattcagcagattttcctgaagggc2040ttcatcgactacctgaacaagaacaatctgaagtatatcgagagcaacaacaacaatgac2100aacaacgacatcttctccaagatcaagatcaaaaaggataacaaagagaagtacgacaag2160atcctgaagaactatgagaagcacaatcggaacaaagaaatccctcacgagatcaatgag2220ttcgtgcgcgagatcaagctggggaagattctgaagtacaccgagaatctgaacatgttt2280tacctgatcctgaagctgctgaaccacaaagagctgaccaacctgaagggcagcctggaa2340aagtaccagtccgccaacaaagaagaaaccttcagcgacgagctggaactgatcaacctg2400ctgaacctggacaacaacagagtgaccgaggacttcgagctggaagccaacgagatcggc2460aagttcctggacttcaacgaaaacaaaatcaaggaccggaaagagctgaaaaagttcgac2520accaacaagatctatttcgacggcgagaacatcatcaagcaccgggccttctacaatatc2580aagaaatacggcatgctgaatctgctggaaaagatcgccgataaggccaagtataagatc2640agcctgaaagaactgaaagagtacagcaacaagaagaatgagattgaaaagaactacacc2700atgcagcagaacctgcaccggaagtacgccagacccaagaaggacgaaaagttcaacgac2760gaggactacaaagagtatgagaaggccatcggcaacatccagaagtacacccacctgaag2820aacaaggtggaattcaatgagctgaacctgctgcagggcctgctgctgaagatcctgcac2880cggctcgtgggctacaccagcatctgggagcgggacctgagattccggctgaagggcgag2940tttcccgagaaccactacatcgaggaaattttcaatttcgacaactccaagaatgtgaag3000tacaaaagcggccagatcgtggaaaagtatatcaacttctacaaagaactgtacaaggac3060aatgtggaaaagcggagcatctactccgacaagaaagtgaagaaactgaagcaggaaaaa3120aaggacctgtacatccggaactacattgcccacttcaactacatcccccacgccgagatt3180agcctgctggaagtgctggaaaacctgcggaagctgctgtcctacgaccggaagctgaag3240aacgccatcatgaagtccatcgtggacattctgaaagaatacggcttcgtggccaccttc3300aagatcggcgctgacaagaagatcgaaatccagaccctggaatcagagaagatcgtgcac3360ctgaagaatctgaagaaaaagaaactgatgaccgaccggaacagcgaggaactgtgcgaa3420ctcgtgaaagtcatgttcgagtacaaggccctggaataa3459<210>24<211>1152<212>prt<213>人工序列(artificialsequence)<400>24metlysvalthrlysvalaspglyileserhislyslystyrileglu151015gluglylysleuvallysserthrserglugluasnargthrserglu202530argleusergluleuleuserileargleuaspiletyrilelysasn354045proaspasnalasergluglugluasnargileargarggluasnleu505560lyslysphepheserasnlysvalleuhisleulysaspservalleu65707580tyrleulysasnarglysglulysasnalavalglnasplysasntyr859095serglugluaspileserglutyraspleulysasnlysasnserphe100105110servalleulyslysileleuleuasngluaspvalasnsergluglu115120125leugluilephearglysaspvalglualalysleuasnlysileasn130135140serleulystyrserpheglugluasnlysalaasntyrglnlysile145150155160asngluasnasnvalglulysvalglyglylysserlysargasnile165170175iletyrasptyrtyrarggluseralalysargasnasptyrileasn180185190asnvalglnglualapheasplysleutyrlyslysgluaspileglu195200205lysleuphepheleuilegluasnserlyslyshisglulystyrlys210215220ileargglutyrtyrhislysileileglyarglysasnasplysglu225230235240asnphealalysileiletyrglugluileglnasnvalasnasnile245250255lysgluleuileglulysileproaspmetsergluleulyslysser260265270glnvalphetyrlystyrtyrleuasplysglugluleuasnasplys275280285asnilelystyralaphecyshisphevalgluileglumetsergln290295300leuleulysasntyrvaltyrlysargleuserasnileserasnasp305310315320lysilelysargilepheglutyrglnasnleulyslysleuileglu325330335asnlysleuleuasnlysleuaspthrtyrvalargasncysglylys340345350tyrasntyrtyrleuglnvalglygluilealathrserasppheile355360365alaargasnargglnasnglualapheleuargasnileileglyval370375380serservalalatyrpheserleuargasnileleugluthrgluasn385390395400gluasnaspilethrglyargmetargglylysthrvallysasnasn405410415lysglygluglulystyrvalserglygluvalasplysiletyrasn420425430gluasnlysglnasngluvallysgluasnleulysmetphetyrser435440445tyrasppheasnmetaspasnlysasngluilegluaspphepheala450455460asnileaspglualaileserserilearghisglyilevalhisphe465470475480asnleugluleugluglylysaspilephealaphelysasnileala485490495prosergluileserlyslysmetpheglnasngluileasnglulys500505510lysleulysleulysilephelysglnleuasnseralaasnvalphe515520525asntyrtyrglulysaspvalileilelystyrleulysasnthrlys530535540pheasnphevalasnlysasnileprophevalproserphethrlys545550555560leutyrasnlysilegluaspleuargasnthrleulysphephetrp565570575servalprolysasplysgluglulysaspalaglniletyrleuleu580585590lysasniletyrtyrglyglupheleuasnlysphevallysasnser595600605lysvalphephelysilethrasngluvalilelysileasnlysgln610615620argasnglnlysthrglyhistyrlystyrglnlysphegluasnile625630635640glulysthrvalprovalglutyrleualaileileglnserargglu645650655metileasnasnglnasplysgluglulysasnthrtyrileaspphe660665670ileglnglnilepheleulysglypheileasptyrleuasnlysasn675680685asnleulystyrilegluserasnasnasnasnaspasnasnaspile690695700pheserlysilelysilelyslysaspasnlysglulystyrasplys705710715720ileleulysasntyrglulyshisasnargasnlysgluileprohis725730735gluileasngluphevalarggluilelysleuglylysileleulys740745750tyrthrgluasnleuasnmetphetyrleuileleulysleuleuasn755760765hislysgluleuthrasnleulysglyserleuglulystyrglnser770775780alaasnlysglugluthrpheseraspgluleugluleuileasnleu785790795800leuasnleuaspasnasnargvalthrgluaspphegluleugluala805810815asngluileglylyspheleuasppheasngluasnlysilelysasp820825830arglysgluleulyslyspheaspthrasnlysiletyrpheaspgly835840845gluasnileilelyshisargalaphetyrasnilelyslystyrgly850855860metleuasnleuleuglulysilealaasplysalalystyrlysile865870875880serleulysgluleulysglutyrserasnlyslysasngluileglu885890895lysasntyrthrmetglnglnasnleuhisarglystyralaargpro900905910lyslysaspglulyspheasnaspgluasptyrlysglutyrglulys915920925alaileglyasnileglnlystyrthrhisleulysasnlysvalglu930935940pheasngluleuasnleuleuglnglyleuleuleulysileleuhis945950955960argleuvalglytyrthrseriletrpgluargaspleuargphearg965970975leulysglyglupheprogluasnhistyrileglugluilepheasn980985990pheaspasnserlysasnvallystyrlysserglyglnilevalglu99510001005lystyrileasnphetyrlysgluleutyrlysaspasnvalglulys101010151020argseriletyrserasplyslysvallyslysleulysglnglulys1025103010351040lysaspleutyrileargasntyrilealahispheasntyrilepro104510501055hisalagluileserleuleugluvalleugluasnleuarglysleu106010651070leusertyrasparglysleulysasnalailemetlysserileval107510801085aspileleulysglutyrglyphevalalathrphelysileglyala109010951100asplyslysilegluileglnthrleugluserglulysilevalhis1105111011151120leulysasnleulyslyslyslysleumetthraspargasnserglu112511301135gluleucysgluleuvallysvalmetpheglutyrlysalaleuglu114011451150<210>25<211>43<212>dna<213>人工序列(artificialsequence)<400>25aattctaatacgactcactataggaatttctactgttgtagat43<210>26<211>32<212>dna<213>人工序列(artificialsequence)<400>26caaagtatgggctacagaaaccgtgccaaaag32<210>27<211>32<212>dna<213>人工序列(artificialsequence)<400>27caaagtatgggcttcagaaaccgtgccaaaag32<210>28<211>32<212>dna<213>人工序列(artificialsequence)<400>28tgggaaaacctatcggaagaaggcaagcctcc32<210>29<211>32<212>dna<213>人工序列(artificialsequence)<400>29tgggaaaacctatcggtagaaggcaagcctcc32<210>30<211>30<212>dna<213>人工序列(artificialsequence)<400>30ggggccaagaaattagagtcctcagaagag30<210>31<211>31<212>dna<213>人工序列(artificialsequence)<400>31ggggccaagaaaattagagtcctcagaagag31<210>32<211>31<212>dna<213>人工序列(artificialsequence)<400>32atgatgaagaaagaggaacgggcttggaaga31<210>33<211>29<212>dna<213>人工序列(artificialsequence)<400>33atgatgaagaaaggaacgggcttggaaga29<210>34<211>33<212>dna<213>人工序列(artificialsequence)<400>34catctcaggtttgttctgagacacctgatgacc33<210>35<211>32<212>dna<213>人工序列(artificialsequence)<400>35catctcaggtttgttctagacacctgatgacc32<210>36<211>33<212>dna<213>人工序列(artificialsequence)<400>36atatacaggatatgcgaattaagaagaaacaaa33<210>37<211>33<212>dna<213>人工序列(artificialsequence)<400>37atatacaggatatgtgaattaagaagaaacaaa33<210>38<211>32<212>dna<213>人工序列(artificialsequence)<400>38taggaggccgagctctgttgcttcgaactcca32<210>39<211>31<212>dna<213>人工序列(artificialsequence)<400>39taggaggccgagctctttgcttcgaactcca31<210>40<211>31<212>dna<213>人工序列(artificialsequence)<400>40tgaggaggtttcgacatggcggtgcagccga31<210>41<211>31<212>dna<213>人工序列(artificialsequence)<400>41tgaggaggtttcgacctggcggtgcagccga31<210>42<211>32<212>dna<213>人工序列(artificialsequence)<400>42aaaaagatcaaagtgctgggctccggtgcgtt32<210>43<211>32<212>dna<213>人工序列(artificialsequence)<400>43aaaaagatcaaagtgctgagctccggtgcgtt32<210>44<211>33<212>dna<213>人工序列(artificialsequence)<400>44atcctctctctgaaatcactgagcaggagaaag33<210>45<211>33<212>dna<213>人工序列(artificialsequence)<400>45atcctctctctgaaatcactgcgcaggagaaag33<210>46<211>1442<212>prt<213>人工序列(artificialsequence)<400>46metglyserserhishishishishishisserserglyleuvalpro151015argglyserhismetalasermetthrglyglyglnglnmetglyarg202530glysergluphegluleuargargglnalacysglyargmetasplys354045lystyrserileglyleuaspileglythrasnservalglytrpala505560valilethraspglutyrlysvalproserlyslysphelysvalleu65707580glyasnthrasparghisserilelyslysasnleuileglyalaleu859095leupheaspserglygluthralaglualathrargleulysargthr100105110alaargargargtyrthrargarglysasnargilecystyrleugln115120125gluilepheserasnglumetalalysvalaspaspserphephehis130135140argleuglugluserpheleuvalglugluasplyslyshisgluarg145150155160hisproilepheglyasnilevalaspgluvalalatyrhisglulys165170175tyrprothriletyrhisleuarglyslysleuvalaspserthrasp180185190lysalaaspleuargleuiletyrleualaleualahismetilelys195200205pheargglyhispheleuilegluglyaspleuasnproaspasnser210215220aspvalasplysleupheileglnleuvalglnthrtyrasnglnleu225230235240pheglugluasnproileasnalaserglyvalaspalalysalaile245250255leuseralaargleuserlysserargargleugluasnleuileala260265270glnleuproglyglulyslysasnglyleupheglyasnleuileala275280285leuserleuglyleuthrproasnphelysserasnpheaspleuala290295300gluaspalalysleuglnleuserlysaspthrtyraspaspaspleu305310315320aspasnleuleualaglnileglyaspglntyralaaspleupheleu325330335alaalalysasnleuseraspalaileleuleuseraspileleuarg340345350valasnthrgluilethrlysalaproleuseralasermetilelys355360365argtyraspgluhishisglnaspleuthrleuleulysalaleuval370375380argglnglnleuproglulystyrlysgluilephepheaspglnser385390395400lysasnglytyralaglytyrileaspglyglyalaserglngluglu405410415phetyrlyspheilelysproileleuglulysmetaspglythrglu420425430gluleuleuvallysleuasnarggluaspleuleuarglysglnarg435440445thrpheaspasnglyserileprohisglnilehisleuglygluleu450455460hisalaileleuargargglngluaspphetyrpropheleulysasp465470475480asnargglulysileglulysileleuthrpheargileprotyrtyr485490495valglyproleualaargglyasnserargphealatrpmetthrarg500505510lysserglugluthrilethrprotrpasnpheglugluvalvalasp515520525lysglyalaseralaglnserpheilegluargmetthrasnpheasp530535540lysasnleuproasnglulysvalleuprolyshisserleuleutyr545550555560glutyrphethrvaltyrasngluleuthrlysvallystyrvalthr565570575gluglymetarglysproalapheleuserglygluglnlyslysala580585590ilevalaspleuleuphelysthrasnarglysvalthrvallysgln595600605leulysgluasptyrphelyslysileglucyspheaspservalglu610615620ileserglyvalgluaspargpheasnalaserleuglythrtyrhis625630635640aspleuleulysileilelysasplysasppheleuaspasngluglu645650655asngluaspileleugluaspilevalleuthrleuthrleupheglu660665670aspargglumetileglugluargleulysthrtyralahisleuphe675680685aspasplysvalmetlysglnleulysargargargtyrthrglytrp690695700glyargleuserarglysleuileasnglyileargasplysglnser705710715720glylysthrileleuasppheleulysseraspglyphealaasnarg725730735asnphemetglnleuilehisaspaspserleuthrphelysgluasp740745750ileglnlysalaglnvalserglyglnglyaspserleuhisgluhis755760765ilealaasnleualaglyserproalailelyslysglyileleugln770775780thrvallysvalvalaspgluleuvallysvalmetglyarghislys785790795800progluasnilevalileglumetalaarggluasnglnthrthrgln805810815lysglyglnlysasnserarggluargmetlysargilegluglugly820825830ilelysgluleuglyserglnileleulysgluhisprovalgluasn835840845thrglnleuglnasnglulysleutyrleutyrtyrleuglnasngly850855860argaspmettyrvalaspglngluleuaspileasnargleuserasp865870875880tyraspvalasphisilevalproglnserpheleulysaspaspser885890895ileaspasnlysvalleuthrargserasplysasnargglylysser900905910aspasnvalproserglugluvalvallyslysmetlysasntyrtrp915920925argglnleuleuasnalalysleuilethrglnarglyspheaspasn930935940leuthrlysalagluargglyglyleusergluleuasplysalagly945950955960pheilelysargglnleuvalgluthrargglnilethrlyshisval965970975alaglnileleuaspserargmetasnthrlystyraspgluasnasp980985990lysleuilearggluvallysvalilethrleulysserlysleuval99510001005seraspphearglysasppheglnphetyrlysvalarggluileasn101010151020asntyrhishisalahisaspalatyrleuasnalavalvalglythr1025103010351040alaleuilelyslystyrprolysleuglusergluphevaltyrgly104510501055asptyrlysvaltyraspvalarglysmetilealalysserglugln106010651070gluileglylysalathralalystyrphephetyrserasnilemet107510801085asnphephelysthrgluilethrleualaasnglygluilearglys109010951100argproleuilegluthrasnglygluthrglygluilevaltrpasp1105111011151120lysglyargaspphealathrvalarglysvalleusermetprogln112511301135valasnilevallyslysthrgluvalglnthrglyglypheserlys114011451150gluserileleuprolysargasnserasplysleuilealaarglys115511601165lysasptrpaspprolyslystyrglyglypheaspserprothrval117011751180alatyrservalleuvalvalalalysvalglulysglylysserlys1185119011951200lysleulysservallysgluleuleuglyilethrilemetgluarg120512101215serserpheglulysasnproileasppheleuglualalysglytyr122012251230lysgluvallyslysaspleuileilelysleuprolystyrserleu123512401245phegluleugluasnglyarglysargmetleualaseralaglyglu125012551260leuglnlysglyasngluleualaleuproserlystyrvalasnphe1265127012751280leutyrleualaserhistyrglulysleulysglyserprogluasp128512901295asngluglnlysglnleuphevalgluglnhislyshistyrleuasp130013051310gluileilegluglnileserglupheserlysargvalileleuala131513201325aspalaasnleuasplysvalleuseralatyrasnlyshisargasp133013351340lysproilearggluglnalagluasnileilehisleuphethrleu1345135013551360thrasnleuglyalaproalaalaphelystyrpheaspthrthrile136513701375asparglysargtyrthrserthrlysgluvalleuaspalathrleu138013851390ilehisglnserilethrglyleutyrgluthrargileaspleuser139514001405glnleuglyglyaspalaalaalaleuaspleuglulysargproala141014151420alathrlyslysalaglyglnalalyslyslysgluhishishishis1425143014351440hishis<210>47<211>20<212>dna<213>人工序列(artificialsequence)<400>47cggagatgtcttgatagcga20<210>48<211>20<212>dna<213>人工序列(artificialsequence)<400>48agcttcctagcaagtaacag20<210>49<211>20<212>dna<213>人工序列(artificialsequence)<400>49gcgcatcttccctttgttat20<210>50<211>20<212>dna<213>人工序列(artificialsequence)<400>50gtccggaagaggaactagaa20<210>51<211>20<212>dna<213>人工序列(artificialsequence)<400>51ctccgtgccgctggtttatt20<210>52<211>20<212>dna<213>人工序列(artificialsequence)<400>52cagaagctctgcattcatcc20<210>53<211>20<212>dna<213>人工序列(artificialsequence)<400>53ggtgatgtgagttctagtcc20<210>54<211>20<212>dna<213>人工序列(artificialsequence)<400>54ggaccgattgtgagaaatgc20<210>55<211>20<212>dna<213>人工序列(artificialsequence)<400>55gtgcccagtgtgatgatatt20<210>56<211>20<212>dna<213>人工序列(artificialsequence)<400>56gcatggcttttggctgttcg20<210>57<211>20<212>dna<213>人工序列(artificialsequence)<400>57gctgtgatttcaacaggacg20<210>58<211>20<212>dna<213>人工序列(artificialsequence)<400>58agaccattggagactacacg20<210>59<211>20<212>dna<213>人工序列(artificialsequence)<400>59atagtactaggctgcctcac20<210>60<211>20<212>dna<213>人工序列(artificialsequence)<400>60catgctccgccacctctacc20<210>61<211>20<212>dna<213>人工序列(artificialsequence)<400>61ctgcagcttcgaagcctcac20<210>62<211>20<212>dna<213>人工序列(artificialsequence)<400>62cttgttgtggtttctcaacc20<210>63<211>20<212>dna<213>人工序列(artificialsequence)<400>63ggaagacgccctcagaagat20<210>64<211>20<212>dna<213>人工序列(artificialsequence)<400>64gcctgtaatcccagctactc20<210>65<211>20<212>dna<213>人工序列(artificialsequence)<400>65caggctagagtgaaatggtg20<210>66<211>20<212>dna<213>人工序列(artificialsequence)<400>66cttccttgtaccaacacgta20<210>67<211>20<212>dna<213>人工序列(artificialsequence)<400>67caggtgtgtaccaacacgta20<210>68<211>20<212>dna<213>人工序列(artificialsequence)<400>68ccaggagtaccagctgccat20<210>69<211>20<212>dna<213>人工序列(artificialsequence)<400>69tcttcacatcatgcagcagc20<210>70<211>20<212>dna<213>人工序列(artificialsequence)<400>70agtggccaggattgattcag20<210>71<211>20<212>dna<213>人工序列(artificialsequence)<400>71gacagcagaatgaccttggt20<210>72<211>20<212>dna<213>人工序列(artificialsequence)<400>72cctgtcttgttcccagtcat20<210>73<211>20<212>dna<213>人工序列(artificialsequence)<400>73gtaagcttacctagagaccc20<210>74<211>20<212>dna<213>人工序列(artificialsequence)<400>74gctgtgatttcaacaggacg20<210>75<211>20<212>dna<213>人工序列(artificialsequence)<400>75ggaccctatctgaatgtgca20<210>76<211>20<212>dna<213>人工序列(artificialsequence)<400>76gtgcattccacgcgtaaaac20<210>77<211>20<212>dna<213>人工序列(artificialsequence)<400>77tgcttattgccaccacctgc20<210>78<211>20<212>dna<213>人工序列(artificialsequence)<400>78cctgcaggaaccctaaaata20<210>79<211>20<212>dna<213>人工序列(artificialsequence)<400>79ccatattttagggttcctgc20<210>80<211>20<212>dna<213>人工序列(artificialsequence)<400>80tgcaggtggtggcaataagc20<210>81<211>20<212>dna<213>人工序列(artificialsequence)<400>81aatctcccccacccttaaga20<210>82<211>20<212>dna<213>人工序列(artificialsequence)<400>82tgacatcaattattatacat20<210>83<211>20<212>dna<213>人工序列(artificialsequence)<400>83gcatttctcagtcctaaaca20<210>84<211>20<212>dna<213>人工序列(artificialsequence)<400>84gggtaaccgtgcggtcgtac20<210>85<211>10<212>dna<213>人工序列(artificialsequence)<400>85gggtaaccgt10<210>86<211>5<212>dna<213>人工序列(artificialsequence)<400>86gggta5<210>87<211>1951<212>prt<213>人工序列(artificialsequence)<400>87asplyslystyrserileglyleuaspileglythrasnservalgly151015trpalavalilethraspglutyrlysvalproserlyslysphelys202530valleuglyasnthrasparghisserilelyslysasnleuilegly354045alaleuleupheaspserglygluthralaglualathrargleulys505560argthralaargargargtyrthrargarglysasnargilecystyr65707580leuglngluilepheserasnglumetalalysvalaspaspserphe859095phehisargleuglugluserpheleuvalglugluasplyslyshis100105110gluarghisproilepheglyasnilevalaspgluvalalatyrhis115120125glulystyrprothriletyrhisleuarglyslysleuvalaspser130135140thrasplysalaaspleuargleuiletyrleualaleualahismet145150155160ilelyspheargglyhispheleuilegluglyaspleuasnproasp165170175asnseraspvalasplysleupheileglnleuvalglnthrtyrasn180185190glnleupheglugluasnproileasnalaserglyvalaspalalys195200205alaileleuseralaargleuserlysserargargleugluasnleu210215220ilealaglnleuproglyglulyslysasnglyleupheglyasnleu225230235240ilealaleuserleuglyleuthrproasnphelysserasnpheasp245250255leualagluaspalalysleuglnleuserlysaspthrtyraspasp260265270aspleuaspasnleuleualaglnileglyaspglntyralaaspleu275280285pheleualaalalysasnleuseraspalaileleuleuseraspile290295300leuargvalasnthrgluilethrlysalaproleuseralasermet305310315320ilelysargtyraspgluhishisglnaspleuthrleuleulysala325330335leuvalargglnglnleuproglulystyrlysgluilephepheasp340345350glnserlysasnglytyralaglytyrileaspglyglyalasergln355360365glugluphetyrlyspheilelysproileleuglulysmetaspgly370375380thrglugluleuleuvallysleuasnarggluaspleuleuarglys385390395400glnargthrpheaspasnglyserileprohisglnilehisleugly405410415gluleuhisalaileleuargargglngluaspphetyrpropheleu420425430lysaspasnargglulysileglulysileleuthrpheargilepro435440445tyrtyrvalglyproleualaargglyasnserargphealatrpmet450455460thrarglysserglugluthrilethrprotrpasnpheglugluval465470475480valasplysglyalaseralaglnserpheilegluargmetthrasn485490495pheasplysasnleuproasnglulysvalleuprolyshisserleu500505510leutyrglutyrphethrvaltyrasngluleuthrlysvallystyr515520525valthrgluglymetarglysproalapheleuserglygluglnlys530535540lysalailevalaspleuleuphelysthrasnarglysvalthrval545550555560lysglnleulysgluasptyrphelyslysileglucyspheaspser565570575valgluileserglyvalgluaspargpheasnalaserleuglythr580585590tyrhisaspleuleulysileilelysasplysasppheleuaspasn595600605glugluasngluaspileleugluaspilevalleuthrleuthrleu610615620phegluaspargglumetileglugluargleulysthrtyralahis625630635640leupheaspasplysvalmetlysglnleulysargargargtyrthr645650655glytrpglyargleuserarglysleuileasnglyileargasplys660665670glnserglylysthrileleuasppheleulysseraspglypheala675680685asnargasnphemetglnleuilehisaspaspserleuthrphelys690695700gluaspileglnlysalaglnvalserglyglnglyaspserleuhis705710715720gluhisilealaasnleualaglyserproalailelyslysglyile725730735leuglnthrvallysvalvalaspgluleuvallysvalmetglyarg740745750hislysprogluasnilevalileglumetalaarggluasnglnthr755760765thrglnlysglyglnlysasnserarggluargmetlysargileglu770775780gluglyilelysgluleuglyserglnileleulysgluhisproval785790795800gluasnthrglnleuglnasnglulysleutyrleutyrtyrleugln805810815asnglyargaspmettyrvalaspglngluleuaspileasnargleu820825830serasptyraspvalasphisilevalproglnserpheleulysasp835840845aspserileaspasnlysvalleuthrargserasplysasnarggly850855860lysseraspasnvalproserglugluvalvallyslysmetlysasn865870875880tyrtrpargglnleuleuasnalalysleuilethrglnarglysphe885890895aspasnleuthrlysalagluargglyglyleusergluleuasplys900905910alaglypheilelysargglnleuvalgluthrargglnilethrlys915920925hisvalalaglnileleuaspserargmetasnthrlystyraspglu930935940asnasplysleuilearggluvallysvalilethrleulysserlys945950955960leuvalseraspphearglysasppheglnphetyrlysvalargglu965970975ileasnasntyrhishisalahisaspalatyrleuasnalavalval980985990glythralaleuilelyslystyrprolysleugluserglupheval99510001005tyrglyasptyrlysvaltyraspvalarglysmetilealalysser101010151020gluglngluileglylysalathralalystyrphephetyrserasn1025103010351040ilemetasnphephelysthrgluilethrleualaasnglygluile104510501055arglysargproleuilegluthrasnglygluthrglygluileval106010651070trpasplysglyargaspphealathrvalarglysvalleusermet107510801085proglnvalasnilevallyslysthrgluvalglnthrglyglyphe109010951100serlysgluserileleuprolysargasnserasplysleuileala1105111011151120arglyslysasptrpaspprolyslystyrglyglypheaspserpro112511301135thrvalalatyrservalleuvalvalalalysvalglulysglylys114011451150serlyslysleulysservallysgluleuleuglyilethrilemet115511601165gluargserserpheglulysasnproileasppheleuglualalys117011751180glytyrlysgluvallyslysaspleuileilelysleuprolystyr1185119011951200serleuphegluleugluasnglyarglysargmetleualaserala120512101215glygluleuglnlysglyasngluleualaleuproserlystyrval122012251230asnpheleutyrleualaserhistyrglulysleulysglyserpro123512401245gluaspasngluglnlysglnleuphevalgluglnhislyshistyr125012551260leuaspgluileilegluglnileserglupheserlysargvalile1265127012751280leualaaspalaasnleuasplysvalleuseralatyrasnlyshis128512901295argasplysproilearggluglnalagluasnileilehisleuphe130013051310thrleuthrasnleuglyalaproalaalaphelystyrpheaspthr131513201325thrileasparglysargtyrthrserthrlysgluvalleuaspala133013351340thrleuilehisglnserilethrglyleutyrgluthrargileasp1345135013551360leuserglnleuglyglyaspalaalaalaleuaspleuglnvallys136513701375glnglnileglnleuargargarggluvalaspgluthralaaspleu138013851390proalagluleuproproleuleuargargleutyralaserarggly139514001405valargseralaglngluleugluargservallysglymetleupro141014151420trpglnglnleuserglyvalglulysalavalgluileleutyrasn1425143014351440alaphearggluglythrargileilevalvalglyasppheaspala144514501455aspglyalathrserthralaleuservalleualametargserleu146014651470glycysserasnileasptyrleuvalproasnargphegluaspgly147514801485tyrglyleuserprogluvalvalaspglnalahisalaargglyala149014951500glnleuilevalthrvalaspasnglyileserserhisalaglyval1505151015151520gluhisalaargserleuglyileprovalilevalthrasphishis152515301535leuproglygluthrleuproalaalaglualaileileasnproasn154015451550leuargaspcysasnpheproserlysserleualaglyvalglyval155515601565alaphetyrleumetleualaleuargthrpheleuargaspglngly157015751580trppheaspgluargglyilealaileproasnleualagluleuleu1585159015951600aspleuvalalaleuglythrvalalaaspvalvalproleuaspala160516101615asnasnargileleuthrtrpglnglymetserargileargalagly162016251630lyscysargproglyilelysalaleuleugluvalalaasnargasp163516401645alaglnlysleualaalaseraspleuglyphealaleuglyproarg165016551660leuasnalaalaglyargleuaspaspmetservalglyvalalaleu1665167016751680leuleucysaspasnileglyglualaargvalleualaasngluleu168516901695aspalaleuasnglnthrarglysgluilegluglnglymetglnval170017051710glualaleuthrleucysglulysleugluargserargaspthrleu171517201725proglyglyleualamettyrhisproglutrphisglnglyvalval173017351740glyileleualaserargilelysgluargphehisargprovalile1745175017551760alaphealaproalaglyaspglythrleulysglyserglyargser176517701775ileglnglyleuhismetargaspalaleugluargleuaspthrleu178017851790tyrproglymetileleulyspheglyglyhisalametalaalagly179518001805leuserleuglugluasplysphegluleupheglnglnargphegly181018151820gluleuvalthrglutrpleuaspproserleuleuglnglygluval1825183018351840valseraspglyproleuserproalaglumetthrmetgluvalala184518501855glnleuleuargaspalaglyprotrpglyglnmetpheproglupro186018651870leupheaspglyhispheargleuleuglnglnargleuvalglyglu187518801885arghisleulysvalmetvalgluprovalglyglyglyproleuleu189018951900aspglyilealapheasnvalaspthralaleutrpproaspasngly1905191019151920valarggluvalglnleualatyrlysleuaspileasngluphearg192519301935glyasnargserleuglnileileileaspasniletrpproile194019451950<210>88<211>96<212>dna<213>人工序列(artificialsequence)<400>88cggagatgtcttgatagcgagttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>89<211>96<212>dna<213>人工序列(artificialsequence)<400>89agcttcctagcaagtaacaggttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>90<211>96<212>dna<213>人工序列(artificialsequence)<400>90gcgcatcttccctttgttatgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>91<211>96<212>dna<213>人工序列(artificialsequence)<400>91gtccggaagaggaactagaagttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>92<211>96<212>dna<213>人工序列(artificialsequence)<400>92ctccgtgccgctggtttattgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>93<211>96<212>dna<213>人工序列(artificialsequence)<400>93cagaagctctgcattcatccgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>94<211>96<212>dna<213>人工序列(artificialsequence)<400>94ggtgatgtgagttctagtccgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>95<211>96<212>dna<213>人工序列(artificialsequence)<400>95ggaccgattgtgagaaatgcgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>96<211>96<212>dna<213>人工序列(artificialsequence)<400>96gtgcccagtgtgatgatattgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>97<211>96<212>dna<213>人工序列(artificialsequence)<400>97gcatggcttttggctgttcggttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>98<211>96<212>dna<213>人工序列(artificialsequence)<400>98gctgtgatttcaacaggacggttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>99<211>96<212>dna<213>人工序列(artificialsequence)<400>99agaccattggagactacacggttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>100<211>96<212>dna<213>人工序列(artificialsequence)<400>100atagtactaggctgcctcacgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>101<211>96<212>dna<213>人工序列(artificialsequence)<400>101catgctccgccacctctaccgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>102<211>96<212>dna<213>人工序列(artificialsequence)<400>102ctgcagcttcgaagcctcacgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>103<211>96<212>dna<213>人工序列(artificialsequence)<400>103cttgttgtggtttctcaaccgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>104<211>96<212>dna<213>人工序列(artificialsequence)<400>104ggaagacgccctcagaagatgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>105<211>96<212>dna<213>人工序列(artificialsequence)<400>105gcctgtaatcccagctactcgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>106<211>96<212>dna<213>人工序列(artificialsequence)<400>106caggctagagtgaaatggtggttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>107<211>96<212>dna<213>人工序列(artificialsequence)<400>107cttccttgtaccaacacgtagttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>108<211>96<212>dna<213>人工序列(artificialsequence)<400>108caggtgtgtaccaacacgtagttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>109<211>96<212>dna<213>人工序列(artificialsequence)<400>109ccaggagtaccagctgccatgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>110<211>96<212>dna<213>人工序列(artificialsequence)<400>110tcttcacatcatgcagcagcgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>111<211>96<212>dna<213>人工序列(artificialsequence)<400>111agtggccaggattgattcaggttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>112<211>96<212>dna<213>人工序列(artificialsequence)<400>112gacagcagaatgaccttggtgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>113<211>96<212>dna<213>人工序列(artificialsequence)<400>113cctgtcttgttcccagtcatgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>114<211>96<212>dna<213>人工序列(artificialsequence)<400>114gtaagcttacctagagacccgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>115<211>96<212>dna<213>人工序列(artificialsequence)<400>115gacaccctaacagggttatggttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>116<211>96<212>dna<213>人工序列(artificialsequence)<400>116ctcactgcagccttgacatcgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>117<211>96<212>dna<213>人工序列(artificialsequence)<400>117cgtccaagtgtgtgagtgaggttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>118<211>96<212>dna<213>人工序列(artificialsequence)<400>118gtggaaacctcagagatgtcgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>119<211>96<212>dna<213>人工序列(artificialsequence)<400>119ggaccctatctgaatgtgcagttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>120<211>96<212>dna<213>人工序列(artificialsequence)<400>120gtgcattccacgcgtaaaacgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>121<211>96<212>dna<213>人工序列(artificialsequence)<400>121tgcttattgccaccacctgcgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>122<211>96<212>dna<213>人工序列(artificialsequence)<400>122cctgcaggaaccctaaaatagttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>123<211>96<212>dna<213>人工序列(artificialsequence)<400>123ccatattttagggttcctgcgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>124<211>96<212>dna<213>人工序列(artificialsequence)<400>124tgcaggtggtggcaataagcgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>125<211>96<212>dna<213>人工序列(artificialsequence)<400>125aatctcccccacccttaagagttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>126<211>96<212>dna<213>人工序列(artificialsequence)<400>126tgacatcaattattatacatgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>127<211>96<212>dna<213>人工序列(artificialsequence)<400>127gcatttctcagtcctaaacagttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>128<211>96<212>dna<213>人工序列(artificialsequence)<400>128gggtaaccgtgcggtcgtacgttttagagctagaaatagcaagttaaaataaggctagtc60cgttatcaacttgaaaaagtggcaccgagtcggtgc96<210>129<211>86<212>dna<213>人工序列(artificialsequence)<400>129gggtaaccgtgttttagagctagaaatagcaagttaaaataaggctagtccgttatcaac60ttgaaaaagtggcaccgagtcggtgc86<210>130<211>81<212>dna<213>人工序列(artificialsequence)<400>130gggtagttttagagctagaaatagcaagttaaaataaggctagtccgttatcaacttgaa60aaagtggcaccgagtcggtgc81<210>131<211>249<212>dna<213>人工序列(artificialsequence)<400>131gagggcctatttcccatgattccttcatatttgcatatacgatacaaggctgttagagag60ataattggaattaatttgactgtaaacacaaagatattagtacaaaatacgtgacgtaga120aagtaataatttcttgggtagtttgcagttttaaaattatgttttaaaatggactatcat180atgcttaccgtaacttgaaagtatttcgatttcttggctttatatatcttgtggaaagga240cgaaacacc249<210>132<211>24<212>rna<213>人工序列(artificialsequence)<400>132cactggcgagcgtcatcttcctac24<210>133<211>1431<212>dna<213>人工序列(artificialsequence)<400>133gtaaaacgacggccagtgaattgtaatacgactcactatagggcgaattgggcccgacgt60cgcatgctcccggccgccatggcggccgcgggaattcgattgtccctttggggactgaga120tgctcctttccagacctggtgcaaggcgtgactcctgctgacaatggggcttcctcagga180cagcactgccctcccacggggttttgctcctatcctcagcttgtccctgcagagctgggc240gtgcccagcaactgcatgcaggcatgccgtgaaggtgtatcaaacgctgatgtgacaggt300gcctcctgccttacagctgaggatgaatctgaacatgtcagagtccagggaatgccctgc360tgggtcccgggaacccagatgtcaacctgagccaggatccatgtcccagacaaatggaag420gactaccccagcaggagggcacgctccccacctgctgtgtcccaaccccagggcaggggc480tgctgacctggaaggtgacccacaaggtatagagctgggcggctttcctcgttataggtg540gagtcttggcccagatgtccgagagccgagcatgtccggtgacgatgtccaccacagggc600cagtcagggcgcagctgtactggtcacaagccatgatgaagtagtagacgatgaaggggg660cgaacagcagtaggaagatgacgctcgccagtgaaaaccagtccacctccctgcgaggac720ggatgcaggcagtcacactggggcccatctgccctgggccccaccaggaccctaagaggc780tctgtgtgggagaactgttgctcaaacccaccagtaccccatgacagaaggcattagctc840ctagcacgggccctccttgcggccagggaagccactcaacatgcctgctctactgtagtt900gattaactggtggcactatctgtggcccccatggaaggcccagagctcagaatatgagcg960gaggtaggtctttcacaaccaccaaggccagtggtttccccagttccaggtcggagagga1020tactcaccctgcacgaagtccccatggttctatggcgaaatgggagctgtgacaataggt1080catacccccagatggtgggaaggccccagctgccctttaagcctttatctttttttctaa1140gttgttgactgaccatgatttctagtttctttttataattccttactttataaagtggaa1200aactgtttacctttcattagtcttgacaaaaaaaaaaaaatcttttttaaaaaggtcctt1260cattcctttgggtccatacaattccaaaggctggaaagaatcactagtgaattcgcggcc1320gcctgcaggtcgaccatatgggagagctcccaacgcgttggatgcatagcttgagtattc1380tatagtgtcacctaaatagcttggcgtaatcatggtcatagctgtttcctg1431<210>134<211>544<212>dna<213>人工序列(artificialsequence)<400>134gagccaagctctccatctagtggacagggaagctagcagcaaaccttcccttcactacaa60aacttcattgcttggccaaaaagagagttaattcaatgtagacatctatgtaggcaatta120aaaacctattgatgtataaaacagtttgcattcatggagggcaactaaatacattctagg180actttataaaagatcactttttatttatgcacagggtggaacaagatggattatcaagtg240tcaagtccaatctatgacatcaattattatacatcggagccctgccaaaaaatcaatgtg300aagcaaatcgcagcccgcctcctgcctccgctctactcactggtgttcatctttggtttt360gtgggcaacatgctggtcatcctcatcctgataaactgcaaaaggctgaagagcatgact420gacatctacctgctcaacctggccatctctgacctgtttttccttcttactgtccccttc480tgggctcactatgctgccgcccagtgggactttggaaatacaatgtgtcaactcttgaca540gggc544<210>135<211>3825<212>dna<213>人工序列(artificialsequence)<400>135atgggcaaaaaccaaaatttccaagaatttatcggagtgagccccctgcagaaaaccctc60cggaacgagcttattccgactgagaccacaaagaaaaatataacccagctggacttgctg120actgaagatgagatccgcgcccagaaccgggaaaagctcaaagagatgatggacgattat180taccgcaatgttattgacagtacccttcacgtcgggatcgctgtggattggtcttatctg240ttcagctgcatgcggaaccatttgcgcgaaaattccaaggagtcaaaacgggaactggag300cgcacacaggacagcattcggagtcagatacacaacaagtttgccgaacgcgcagatttc360aaagacatgtttggcgcctctatcattaccaagctccttcctacttacatcaaacaaaat420agcgagtattccgaacggtacgatgagtcaatggaaattctgaagttgtatggtaaattc480accacaagcctgaccgactactttgagactcgcaagaacatattcagtaaagaaaagatc540tctagcgctgtaggctatcggattgtggaggaaaatgccgagatctttctccagaaccag600aatgcatacgatcgcatttgtaaaatagccggacttgacctgcatgggttggataacgaa660atcaccgcttatgttgacggcaagacactgaaagaggtctgctccgatgaaggtttcgcc720aaggcaattacccaagagggcatcgatcggtacaatgaagccattggagctgtgaaccag780tatatgaatctcctttgtcagaaaaacaaggccctgaaacccgggcaatttaagatgaaa840cgcttgcacaagcagatactgtgcaaaggcactacctcattcgatatcccgaagaaattt900gagaatgacaagcaggtatacgatgcagtgaacagcttcacagaaattgttaccaaaaat960aacgacctcaagcggcttctgaatatcactcaaaacgccaatgattatgacatgaacaaa1020atttacgtcgtggctgatgcctatagtatgatatctcagtttatcagcaagaaatggaat1080ttgattgaggaatgtctgctcgactactattccgataaccttccaggtaagggcaatgca1140aaagagaacaaggtaaaaaaggccgtgaaagaagagacctaccgctcagttagccagctg1200aatgaagtcatcgagaagtattacgtggaaaaaacaggacaaagtgtatggaaggtggag1260tcttatattagctccttggctgaaatgataaaactggagctctgccatgaaatcgacaac1320gatgagaagcacaatcttattgaggacgatgagaaaatctcagaaattaaggagctgttg1380gacatgtacatggatgttttccatataatcaaagtctttcgggtgaacgaagtactgaat1440ttcgacgagaccttttatagcgaaatggatgagatttaccaggacatgcaggaaatcgtg1500cccctctataaccacgttcgcaattacgtcactcaaaagccgtataaacaggagaagtac1560cggctttatttccatacccctacactggccaacgggtggagtaaatctaaggaatacgat1620aataacgcaattatattggtgcgcgaggacaaatattacctgggcatcctcaatgccaag1680aaaaagcccagcaaagaaattatggctggtaaggaggattgttccgaacacgcctatgca1740aaaatgaactactatcttctgccgggcgccaataagatgttgccaaaagtatttctgtca1800aagaaaggaatccaggactaccatcccagcagttatattgtggaggggtacaacgaaaag1860aaacacataaagggctctaaaaatttcgatatccggttttgccgcgacctcattgattat1920ttcaaggagtgtatcaaaaagcatccggactggaacaaatttaatttcgaatttagcgct1980accgagacttacgaagatatttccgttttctatcgggaggtcgaaaagcaaggttaccgc2040gtggagtggacctatataaactcagaggacatccagaaacttgaggaagatggccagctg2100tttttgttccaaatttacaataaggactttgccgtaggaagcacagggaaacctaacctg2160cacaccctctatcttaagaatctgttcagtgaggaaaacttgcgggatatcgtgctgaaa2220ctcaatggcgaggcagaaatttttttccgcaagtctagcgttcagaaacccgtcatacat2280aagtgcggttccatccttgtgaaccggacttacgagattaccgaatcaggcacaacccgc2340gtacagagcatcccggagagtgaatatatggagctgtatcggtattttaattctgaaaaa2400caaattgagttgagcgacgaagccaagaaatacctggataaggtgcagtgtaacaaagct2460aagactgacatagttaaagattatcgctacaccatggacaagttctttatccacctccca2520attacaatcaatttcaaagtcgataagggaaacaatgtgaacgccattgcacagcaatat2580atagccgggcggaaagaccttcatgtaatcggcattgatcgcggtgagcggaatctgatc2640tacgtgtccgttattgacatgtatggccgcatattggaacagaagtcatttaacctggtc2700gagcaggtgagcagtcaaggaaccaaacggtactatgattacaaggaaaaactccagaat2760cgcgaggaagagcgggacaaggctcgccagtcttggaaaactatcgggaagattaaagaa2820cttaaggagggctatctgagctccgtaatccacgaaattgcccaaatggtggttaaatac2880aacgcaataatcgccatggaggatttgaattatggtttcaagcggggccgctttaaagtc2940gaacggcaggtgtaccagaagttcgagaccatgctgatttcaaaactcaactatcttgct3000gacaagagccaagccgtagatgaacccggagggattctgcgcggctaccagatgacatat3060gtgccggacaatattaaaaacgttggtcggcagtgcggcataatcttttacgtccctgca3120gcctataccagtaagattgatcccactaccggattcatcaatgcttttaaacgcgacgtg3180gtatctacaaacgatgccaaggagaatttcttgatgaaatttgacagcattcaatacgat3240atagaaaaggggctgttcaaattttccttcgactataagaactttgcaacccataaactc3300actcttgccaagaccaaatgggatgtgtacacaaatggcacccggattcagaacatgaag3360gttgagggtcactggctgtcaatggaagtcgagttgactaccaaaatgaaggaactgctc3420gacgatagccatattccgtatgaggaaggccagaatatccttgacgatctgcgcgagatg3480aaagacattacaaccatagtgaacggaatcttggaaattttctggctgactgtacaactc3540cggaatagtcgcatcgataacccagactacgatcggattatatctcccgtgcttaataag3600aacggggagtttttcgacagcgatgaatataattcctacatcgacgctcagaaagccccg3660ctgcctattgatgcagacgccaacggcgctttttgtatcgccttgaagggtatgtatacc3720gcaaatcagattaaagagaactgggttgaaggcgagaagctgcccgccgattgcctcaaa3780atagaacacgcttcatggcttgccttcatgcaaggagagcgcggg3825<210>136<211>1275<212>prt<213>人工序列(artificialsequence)<400>136metglylysasnglnasnpheglnglupheileglyvalserproleu151015glnlysthrleuargasngluleuileprothrgluthrthrlyslys202530asnilethrglnleuaspleuleuthrgluaspgluileargalagln354045asnargglulysleulysglumetmetaspasptyrtyrargasnval505560ileaspserthrleuhisvalglyilealavalasptrpsertyrleu65707580phesercysmetargasnhisleuarggluasnserlysgluserlys859095arggluleugluargthrglnaspserileargserglnilehisasn100105110lysphealagluargalaaspphelysaspmetpheglyalaserile115120125ilethrlysleuleuprothrtyrilelysglnasnserglutyrser130135140gluargtyraspglusermetgluileleulysleutyrglylysphe145150155160thrthrserleuthrasptyrphegluthrarglysasnilepheser165170175lysglulysileserseralavalglytyrargilevalglugluasn180185190alagluilepheleuglnasnglnasnalatyraspargilecyslys195200205ilealaglyleuaspleuhisglyleuaspasngluilethralatyr210215220valaspglylysthrleulysgluvalcysseraspgluglypheala225230235240lysalailethrglngluglyileaspargtyrasnglualailegly245250255alavalasnglntyrmetasnleuleucysglnlysasnlysalaleu260265270lysproglyglnphelysmetlysargleuhislysglnileleucys275280285lysglythrthrserpheaspileprolyslysphegluasnasplys290295300glnvaltyraspalavalasnserphethrgluilevalthrlysasn305310315320asnaspleulysargleuleuasnilethrglnasnalaasnasptyr325330335aspmetasnlysiletyrvalvalalaaspalatyrsermetileser340345350glnpheileserlyslystrpasnleuilegluglucysleuleuasp355360365tyrtyrseraspasnleuproglylysglyasnalalysgluasnlys370375380vallyslysalavallysglugluthrtyrargservalserglnleu385390395400asngluvalileglulystyrtyrvalglulysthrglyglnserval405410415trplysvalglusertyrileserserleualaglumetilelysleu420425430gluleucyshisgluileaspasnaspglulyshisasnleuileglu435440445aspaspglulysilesergluilelysgluleuleuaspmettyrmet450455460aspvalphehisileilelysvalpheargvalasngluvalleuasn465470475480pheaspgluthrphetyrserglumetaspgluiletyrglnaspmet485490495glngluilevalproleutyrasnhisvalargasntyrvalthrgln500505510lysprotyrlysglnglulystyrargleutyrphehisthrprothr515520525leualaasnglytrpserlysserlysglutyraspasnasnalaile530535540ileleuvalarggluasplystyrtyrleuglyileleuasnalalys545550555560lyslysproserlysgluilemetalaglylysgluaspcysserglu565570575hisalatyralalysmetasntyrtyrleuleuproglyalaasnlys580585590metleuprolysvalpheleuserlyslysglyileglnasptyrhis595600605prosersertyrilevalgluglytyrasnglulyslyshisilelys610615620glyserlysasnpheaspileargphecysargaspleuileasptyr625630635640phelysglucysilelyslyshisproasptrpasnlyspheasnphe645650655glupheseralathrgluthrtyrgluaspileservalphetyrarg660665670gluvalglulysglnglytyrargvalglutrpthrtyrileasnser675680685gluaspileglnlysleuglugluaspglyglnleupheleuphegln690695700iletyrasnlysaspphealavalglyserthrglylysproasnleu705710715720histhrleutyrleulysasnleupheserglugluasnleuargasp725730735ilevalleulysleuasnglyglualagluilephephearglysser740745750servalglnlysprovalilehislyscysglyserileleuvalasn755760765argthrtyrgluilethrgluserglythrthrargvalglnserile770775780progluserglutyrmetgluleutyrargtyrpheasnserglulys785790795800glnilegluleuseraspglualalyslystyrleuasplysvalgln805810815cysasnlysalalysthraspilevallysasptyrargtyrthrmet820825830asplysphepheilehisleuproilethrileasnphelysvalasp835840845lysglyasnasnvalasnalailealaglnglntyrilealaglyarg850855860lysaspleuhisvalileglyileaspargglygluargasnleuile865870875880tyrvalservalileaspmettyrglyargileleugluglnlysser885890895pheasnleuvalgluglnvalserserglnglythrlysargtyrtyr900905910asptyrlysglulysleuglnasnarggluglugluargasplysala915920925argglnsertrplysthrileglylysilelysgluleulysglugly930935940tyrleuserservalilehisgluilealaglnmetvalvallystyr945950955960asnalaileilealametgluaspleuasntyrglyphelysarggly965970975argphelysvalgluargglnvaltyrglnlysphegluthrmetleu980985990ileserlysleuasntyrleualaasplysserglnalavalaspglu99510001005proglyglyileleuargglytyrglnmetthrtyrvalproaspasn101010151020ilelysasnvalglyargglncysglyileilephetyrvalproala1025103010351040alatyrthrserlysileaspprothrthrglypheileasnalaphe104510501055lysargaspvalvalserthrasnaspalalysgluasnpheleumet106010651070lyspheaspserileglntyraspileglulysglyleuphelysphe107510801085serpheasptyrlysasnphealathrhislysleuthrleualalys109010951100thrlystrpaspvaltyrthrasnglythrargileglnasnmetlys1105111011151120valgluglyhistrpleusermetgluvalgluleuthrthrlysmet112511301135lysgluleuleuaspaspserhisileprotyrglugluglyglnasn114011451150ileleuaspaspleuargglumetlysaspilethrthrilevalasn115511601165glyileleugluilephetrpleuthrvalglnleuargasnserarg117011751180ileaspasnproasptyraspargileileserprovalleuasnlys1185119011951200asnglygluphepheaspseraspglutyrasnsertyrileaspala120512101215glnlysalaproleuproileaspalaaspalaasnglyalaphecys122012251230ilealaleulysglymettyrthralaasnglnilelysgluasntrp123512401245valgluglyglulysleuproalaaspcysleulysilegluhisala125012551260sertrpleualaphemetglnglygluarggly126512701275<210>137<211>24<212>rna<213>人工序列(artificialsequence)<400>137ggagatgttgcttctcttaattcc24<210>138<211>24<212>rna<213>人工序列(artificialsequence)<400>138tgcacagggtggaacaagatggat24<210>139<211>24<212>rna<213>人工序列(artificialsequence)<400>139ggagatgtcttgatagcgacggga24当前第1页12
相关技术
网友询问留言
已有0条留言
- 还没有人留言评论。精彩留言会获得点赞!
1