一组生物合成创新霉素或开环创新霉素的基因簇的制作方法
本发明属于医药技术领域,具体而言,涉及一组合成创新霉素的基因簇。
背景技术:
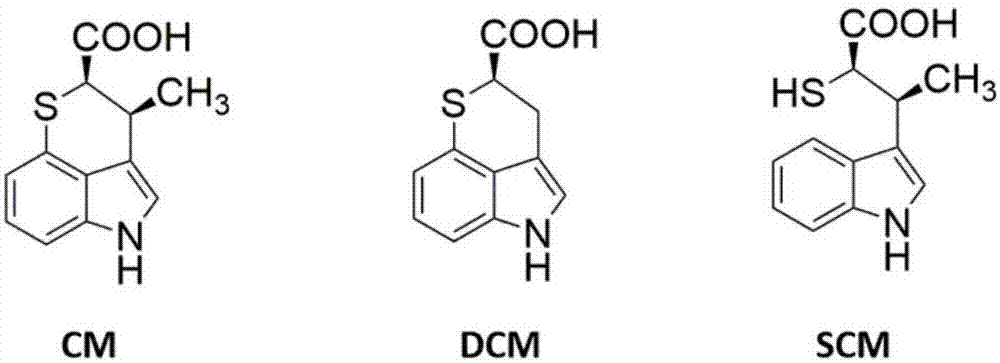
创新霉素(chuangxinmycin,简称cm,化学结构见图1)是20世纪70年代中国医学科学院医药生物技术研究所(原名:中国医学科学院抗菌素研究所)首先发现的一个具有含氮、含硫杂环新骨架的抗生素,由从我国山东济南土壤中分离的1株游动放线菌(actinoplanestsinanensiscpcc200056)产生。在临床上,cm对大肠杆菌等细菌感染所引起的败血症、尿路感染、胆道感染和婴儿腹泻等有一定疗效,临床试用的有效率达77.86%[1]。
由于cm具有新颖的骨架结构和良好的抑菌活性,国内外学者对其进行了多方面研究。在化学方面,已确定cm的绝对构型[2]并建立了cm化学全合成路线,获得了一系列cm衍生物[3-5];在生物学活性方面,已证实cm的抗菌作用是通过选择性抑制色氨酸trna合成酶活性实现的[6];在生物合成方面,戚天庆等从cm产生菌中分离纯化了一种可能参与催化cm生物合成的吲哚丙酮酸甲基转移酶[7],左利杰等从cm产生菌中分离得到3-去甲创新霉素(demethyl-chuangxinmycin,dcm,图1),但目前还不清楚dcm是cm生物合成过程中出现的中间产物还是支路产物[8];许津等利用同位素标记实验推测cm分子中的硫原子可能来源于半胱氨酸(cys)的巯基[9],以及周锡漳等探讨了维生素b12在cm生物合成中的作用[10]。但是到目前为止,cm的生物合成基因簇尚未报道,关于cm中硫原子的掺入机制以及c-s键(碳硫键)的形成等生物合成机制仍为未知。因此,cm生物合成基因簇的解析将阐明其生物合成机制及调控机制,为利用遗传操作提高其产量,或者获得新结构衍生物用于新药发现奠定基础。
众所周知,在微生物次级代谢产物的生物合成基因簇中,其抗性基因和生物合成基因往往共同存在以保护产生菌免于其产生的抗生素的损害。吲哚霉素(indolmycin)和cm的抗菌机制相同,也属于色氨酸trna合成酶抑制剂。2015年,du等在吲哚霉素产生菌全基因组范围内扫描色氨酸trna合成酶的同源基因从而发现吲哚霉素的生物合成基因簇[11]。我们推测,在cm生物合成基因簇内可能存在色氨酸trna合成酶基因。
另一方面,创新霉素结构中一个显著特征是其含有s元素,s的掺入以及c-s键的形成应为其生物合成的主要步骤。到目前为止,次级代谢产物生物合成过程中s的掺入机制仍所知甚少。2014年,sasaki等报道了amycolatopsisorientalissubsp.vineariaba-07585中次级代谢产物be-7585a生物合成中s的掺入机制[12]:利用原核生物初级代谢中维生素b1(thiamine)、钼蝶呤、半胱氨酸/甲硫氨酸等的硫原子掺入机制,即硫载体蛋白(sulfur-carrierprotein,scp)、scp活化蛋白等共同完成。在be-7585a生物合成基因簇内存在催化维生素b1中噻唑合成酶thig[13]的同源蛋白bexx,但该基因簇中不含有scp;在基因组中存在几个scp的同源蛋白(this,moad,cyso和moad2),但仅有唯一的scp活化蛋白thif的同源蛋白moez(c-端还含有硫氰酸酶结构域,催化活化的scp的硫醇化)。sasaki等通过实验证实a.orientalis中此三个元件(bexx,scp,moez)负责be-7585a中2-硫糖结构的s的掺入。
技术实现要素:
本发明首先涉及创新霉素生物合成的基因簇,所述的基因簇依次包含cxna、cxnb、cxnc、cxnd、cxne、cxnf、cxnt、cxnr、trprs九个关键功能基因,以及在创新霉素分子中引入s元素所必需的thig、moez基因,九个关键基因和thig、moez基因的核苷酸序列分别为如seqidno.2~12所示,包含cxna、cxnb、cxnc、cxnd、cxne、cxnf、cxnt、cxnr、trprs九个关键功能基因的基因片段序列如seqidno.1所示,
本发明还涉及创新霉素生物合成基因簇中的各个功能蛋白:
cxna为依赖于维生素b12的自由基s-腺苷甲硫氨酸家族的c-甲基转移酶,其功能为催化吲哚丙酮酸甲基化生成3-甲基吲哚丙酮酸;
cxnb为氨基转移酶,其功能为将创新霉素的生物合成前体色氨酸(l-trp)中的氨基转变为酮基,形成吲哚丙酮酸;
cxnc为还原酶,其功能为将3-甲基吲哚丙酮酸的酮基还原为羟基,形成3-吲哚-2-羟基丁酸;
cxnd为细胞色素p450酶,其功能为催化cm的开环结构(secochuangxinmycin,scm)中的s原子和c原子形成c-s键,形成cm;
cxne为硫载体蛋白,在jamm金属蛋白酶家族的cxnf的作用下,c-末端10个氨基酸残基被水解露出双甘氨酸基序,而后先在scp活化蛋白moez(n-端为类似于scp活化蛋白thif的结构域)作用下生成腺苷酰化的scp,再在moez的c-端硫氰酸酶结构域的作用下形成硫醇化的scp,即为硫原子的供体;
cxnf为jamm金属蛋白酶家族成员,其功能为酶切cxne的c-末端10个氨基酸残基,露出双甘氨酸基序;
cxnr为转录调控蛋白,其功能为调节所述基因簇的表达;
cxnt为创新霉素的转运蛋白;
trprs为色氨酸trna合成酶,是创新霉素的自身抗性基因;
thig为噻唑合成酶,其功能为将硫醇化scp中的s原子转移至还原酶cxnc催化形成的3-吲哚-2-羟基丁酸的羟基,得到3-吲哚-2-巯基丁酸(即cxnd的底物,开环创新霉素);
moez为scp活化蛋白,其作用为活化scp蛋白cxne,使其硫醇化形成硫代羧酸酯。
本发明还涉及所述的基因簇中的各个基因thig、cxna、cxnb、cxnc、cxnd、cxne、cxnf、cxnt、cxnr、trprs、moez编码的蛋白质,其氨基酸序列分别为seqidno.13~23所示。
本发明还涉及一种开环创新霉素scm,其结构式如下式所示
本发明还涉及所述的开环创新霉素scm的生物合成基因簇,包括cxna、cxnb、cxnc、cxne、cxnf、cxnt、cxnr、trprs及thig、moez十个功能基因。
本发明还涉及包括所述的开环创新霉素或创新霉素基因簇的重组载体,优选的,所述的重组载体为包含酵母菌元件(arsh4/cen6复制子和trp1筛选标记)、大肠杆菌元件(pucori复制子)和链霉菌元件(φc31整合酶及其整合位点attp、dna接合转移起始位点orit)的重组载体,最优选的,所述的重组载体为pcap01。
本发明还涉及转化了所述的重组载体的宿主,优选的,所述的宿主为酵母、大肠杆菌或链霉菌,最优选的,所述的宿主为链霉菌。
本发明还涉及通过生物发酵生产cm或scm的方法,其步骤包括:
(1)将所述cm或scm的生物合成基因簇克隆至目标宿主,
(2)发酵所述目标宿主并从发酵液中提取并纯化所述cm或scm。
本发明还涉及所述的创新霉素生物合成基因簇或所述的开环创新霉素生物合成基因簇和/或其编码蛋白在催化合成创新霉素、开环创新霉素或其类似物中的应用。
本发明还涉及所述的cxnr基因或其编码的cxnr蛋白在合成创新霉素、开环创新霉素中的应用。
本发明还涉及一种提高创新霉素、开环创新霉素产量的方法,其特征在于,在生产菌株中过表达所述cxnr基因,所述的过表达的方法为,将高表达所述cxnr基因的重组载体转入创新霉素、开环创新霉素的生产菌株中,优选的,所述的重组载体为质粒pset152,所述的生产菌株为来自中国药学微生物菌种保藏管理中心的野生型菌株actinoplanestsinanensiscpcc200056(chinapharmaceuticalculturecollection)。
本发明还涉及所述的cxnd基因或其编码的cxnd蛋白在以开环创新霉素为底物合成创新霉素中的应用。
本发明还涉及一种合成创新霉素的方法,其特征在于,以开环创新霉素为底物,发酵转化了所述cxnd基因的宿主,所述的宿主不包含所述创新霉素合成基因簇的其他基因,优选的,所述的宿主为链霉菌。
附图说明
图1、创新霉素(cm)、去甲创新霉素(dcm)及开环创新霉素(scm)的结构示意图。
图2、创新霉素生物合成基因簇及其相邻基因:thig(ats3059)与moez(ats6133)位于基因簇外,但推测参与cm的生物合成;左臂和右臂为dnaassembler方法中的捕捉臂;“deleted”为阻断菌株m1146/pcap-cm(δcxna~f)中删除基因部分。
图3、pl-cxnr的质粒图和酶切鉴定结果:
3a、pl-cxnr质粒示意图;
3b、pl-cxnr质粒酶切鉴定结果:lane1~4:ndei和xbai双酶切鉴定pl-cxnr,预计大小6kb和0.95kb;lanem:1kbplusdnaladder(10,000,8,000,6,000,5,000,4,000,3,000,2,000,1,500,1000,800,500,300bp)。
图4、重组菌株的pcr验证:lanem:1kbplusdnaladder(10,000,8,000,6,000,5,000,4,000,3,000,2,000,1,500,1000,800,500,300bp);wt:cpcc200056;-:pcr的阴性对照。
图5、tlc检测各菌株中cm的产量:1~4:200056/pset152的4个单克隆;5~8:200056/pl-cxnr的4个单克隆;wt:cpcc200056;cm:创新霉素标准品。
图6、hplc检测各菌株中cm的产量:
6a、200056/pset152和200056/pl-cxnr其中一个单克隆发酵产物的hplc图;
6b、cm产量的hplc定量比较(200056/pset152与200056/pl-cxnr分别为4个和8个单克隆的统计结果)。
图7、利用dnaassembler构建质粒pcap01-cm及相关验证:
7a、pcap01-cm的构建示意图;
7b、从e.colidh5α中提取的质粒pcap01-cm进行pcr验证。lane1~2:扩增左臂片段,预计大小2265bp;lane3~4:扩增右臂片段,预计大小2225bp;lane5~6:扩增片段1,预计大小4003bp;lane7~8:扩增片段2,预计大小4101bp;lane9~10:扩增片段3,预计大小4049bp;lane11~12:扩增片段4,预计大小4388bp;lanem:1kbplusdnaladder(10,000,8,000,6,000,5,000,4,000,3,000,2,000,1,500,1000,800,500,300bp);
7c、从e.colidh5α中提取的质粒pcap01-cm进行限制性内切酶验证。lane1~2:spei酶切(28242bp);lane3~4:ndei酶切(28242bp);lane5~6:ecori酶切(23132,4336bp);lane7~8:kpni酶切(10834,10483,3340,1884,1601bp);lane9~10:xhoi酶切(12553,6319,3031,2415,1002,951,714,444,399,237,141bp);lanem:1kbdnaladder(10,000,8,000,6,000,5,000,4,000,3,000,2,000,1,000bp)。
图8、重组菌株的pcr验证:
8a、利用φc31整合位点通用引物(pset152与attb_streptomyces)进行验证。预计大小1.6kb;lanem:1kbplusdnaladder(10,000,8,000,6,000,5,000,4,000,3,000,2,000,1,500,1000,800,500,300bp);lane1,13,14:s.coelicolorm1146;lane2~6:m1146/pcap-cm(δcxnd)的5个单克隆;lane7~11:m1146/pset152的5个单克隆;lane12:pcr阴性对照;lane15~19:m1146/pcap-cm(δcxna~f)的5个单克隆;
8b、利用在删除序列的上下游设计的引物(cm-23k-1和cm-23k-2)进行验证。lanem:1kbdnaladder(10,000,8,000,6,000,5,000,4,000,3,000,2,000,1,000bp);lane1:pcr阴性对照;lane2:s.coelicolorm1146;lane3~7:m1146/pcap-cm的5个单克隆,预计大小5,851bp;lane8~15:m1146/pcap-cm(δcxna~f)的8个单克隆,预计大小741bp;
8c、利用φbt1整合位点设计引物(bt1-m1146-f1与bt1-m1146-r1)进行验证。预计大小1.0kb;lanem:1kbplusdnaladder(10,000,8,000,6,000,5,000,4,000,3,000,2,000,1,500,1000,800,500,300bp);lane1~8:m1146/pcap-cm(δcxnd)/pij10500的8个单克隆;lane9~20:m1146/pcap-cm的12个单克隆;lane21:s.coelicolorm1146;lane22:m1146/pcap-cm(δcxnd)。
图9、lc-ms分析异源表达产物:
9a、各菌株发酵产物的hplc分析;i、野生型菌株发酵产物的hplc分析;ii、m1146/pset152发酵产物的hplc分析;iii、m1146/pcap-cm发酵产物的hplc分析;iv、m1146/pcap-cm(δcxna~f)发酵产物的hplc分析;v、m1146/pcap-cm(δcxnd)发酵产物的hplc分析;
9b、图a中各个峰的ms分析图;依次为1(即cm)的ms分析图、2(即dcm)的ms分析图、3(即scm)的ms分析图;c、cm、dcm和scm的紫外吸收光谱;
9c、cm、dcm和scm的紫外吸收光谱。
图10、cm与scm的二级高分辨质谱图:
10a、创新霉素的二级高分辨质谱图;
10b、开环创新霉素的二级高分辨质谱图。
图11、cm生物合成过程示意图:
11a、硫载体蛋白的活化;
11b、cm生物合成途径分析。
具体实施方式
材料与方法
1、菌株、质粒和培养方法
野生型菌株actinoplanestsinanensiscpcc200056来自中国药学微生物菌种保藏管理中心(chinapharmaceuticalculturecollection),其培养和固体发酵均使用isp2培养基(bdno.277010),接合转移使用threemedium65培养基[23];streptomycescoelicolorm1146及其相关链霉菌菌株的培养和接合转移均使用ms培养基[24],固体发酵使用isp2培养基;培养和发酵均于28℃恒温培养7d;所有菌株提取基因组dna时都使用液体φ培养基[25]培养。saccharomycescerevisiaevl6-48是用来克隆生物合成基因簇的宿主菌,使用ypad培养基培养[20],利用dnaassembler进行克隆时使用sd-trp培养基(sigmano.630411和630413)进行筛选,培养温度30℃。escherichiacolidh5α作为通用的大肠杆菌克隆宿主[26],e.coliet12567/puz8002是用于在大肠杆菌及链霉菌之间进行接合转移的宿主[27],二者使用lb培养基37℃恒温培养。当需要用抗生素时,它们的工作浓度如下:阿普霉素(apramycin,am,50μg/ml),潮霉素(hygromycinb,hyg,200μg/ml),卡那霉素(kanamycin,km,50μg/ml),氯霉素(chloramphenicol,cm,30μg/ml)和萘啶酮酸(nalidixicacid,nd,25μg/ml)。本文中所有的菌株和质粒见表1,引物见表2。
表1、菌株和质粒
amr,apramycinresistance;kmr,kanamycinresistance;cmr,chloramphenicolresistance;hygr,hygromycinbresistance
表2、引物
restrictionendonucleaserecognitionsequencesintroducedbytheoligonucleotidesarebold.
2、提取高质量的actinoplanestsinanensiscpcc200056基因组
将a.tsinanensiscpcc200056接种于100ml的φ培养基中,28℃,230rpm振荡培养24h。5,000rpm离心10min收集15ml菌体,并用ste缓冲液(10mmtris-hcl,1mmedta,ph8.0)洗涤菌体一次。用5mlste缓冲液重悬菌体,加入终浓度为5mg/ml的溶菌酶,在37℃恒温水浴中孵育30min。然后加2ml2%sds轻轻混匀后,加入等体积的酚:氯仿:异戊醇(25:24:1),在室温下轻轻摇匀10min,5,000rpm,4℃离心10min。将上清液转移到新的离心管中。加rnasea至终浓度15μg/ml,在37℃恒温水浴中孵育30min,然后加入等体积的酚:氯仿:异戊醇(25:24:1),在室温下轻轻摇匀10min,5,000rpm,4℃离心10min。将上清液转移到新的离心管中。重复抽提1~2次,直至界面没有白色变性蛋白的存在。加入等体积的氯仿抽提一次以除去残留的酚,5,000rpm,4℃离心10min。
将上清液转移到新的离心管中,加入1/20体积的5mnacl和等体积异丙醇,轻轻地充分混匀至dna沉淀。用灭菌的玻璃棒缠绕dna,将其移至新的离心管中,70%的乙醇洗涤一至两次,室温晾干后溶于500μlddh2o,保存于4℃备用。经电泳检测符合送样要求后,送样测序。
3、菌株发酵及产物的tlc、hplc和lc-ms分析
本文中所有菌株都采用isp2培养基进行固体发酵,在28℃恒温摇床间中倒置培养7d,然后将每个isp2平板培养物切碎成1cm×1cm的小方块,用2倍体积乙酸乙酯萃取48h,最后将乙酸乙酯萃取液浓缩至相同体积(250~500μl),获得固体发酵产物。
硅胶板tlc分析的点样量为5μl,展开剂系统为乙酸乙酯-正己烷-二氯甲烷-冰醋酸,9:7:6:0.2,v/v,分别在254和365nm观察。
将最终的乙酸乙酯萃取液过滤,滤膜为0.22μm有机相专用,滤液进行hplc分析。hplc条件包括:1、xselectcshtmc18色谱柱(4.6×150mm,5μm,waters,ireland)或eclipseplusc18色谱柱(4.6×150mm,5μm,agilent,america)或capcellpakadme色谱柱(4.6×250mm,5μm,shiseido,japan);2、流动相a:0.1%ch3cooh,流动相b:乙腈;3、梯度洗脱:在30min内,0.1%ch3cooh-mecn从85(a):15(b)逐渐变换到0(a):100(b);4、流速设定为1.0ml/min;5、检测波长设定为254nm;6、柱温和检测池温度均为室温(25℃)。
进行lc-ms分析时,hplc条件相同,将流速改为0.8ml/min。使用的仪器为1290(lc)-1956singlequadrupolems(agilent,america)或1100(lc)-6300msdtrapms(agilent,america);ms条件为:电喷雾(esi)离子源,负离子全扫描模式,电喷雾电压:4.5kv;加热毛细管温度:325℃;载气:n2。
将化学合成的cm标准品和利用hplc制备纯化的scm进行hresims和hresims-ms分析,使用的仪器为qstartmelitelc/ms/mssystem(appliedbiosystems/msdsciex,singapore),配有esi源,采用负离子tof扫描模式。
4、actinoplanestsinanensiscpcc200056与e.coliet12567/puz8002之间的接合转移
借鉴文献报道的actinoplanesfriuliensis的接合转移方法[23]建立a.tsinanensiscpcc200056的遗传操作系统。a.tsinanensiscpcc200056在isp2平板上于28℃培养7d,然后将培养好的菌体铲下一个大小约1cm×1cm的菌块接种至100mltsb液体培养基中,28℃,220rpm,振荡培养约96h;然后以10%转接至50mltsb液体培养基中,28℃,220rpm,振荡培养约16h;再以20%转接至50mltsb液体培养基中,28℃,220rpm,振荡培养约1~5h,然后离心收集菌丝体(5,000×g,10min),重悬于8~10mltsb中。过夜培养的15ml大肠杆菌供体菌用15mllb洗涤2次,最后重悬于1~2mltsb中。将200μl大肠杆菌供体菌与200μlactinoplanestsinanensiscpcc200056的菌丝体均匀混合后(二者的浓度均接近于108cells/ml),涂布在含有20mmmgcl2的threemedium65平板[23],放入28℃恒温摇床间中倒置培养约16~18h后,向每块平板表面覆盖3ml含有萘啶酸(nd,终浓度为25μg/ml)和阿普霉素(am,终浓度为50μg/ml)的水,在超净台内吹干后放入28℃继续培养,大约3~5d以后挑取接合子,在isp2(含50μg/mlam)平板划线扩大培养,约7d后将菌株接种于5ml的φ培养基,28℃,220rpm振荡培养24~48h,利用magen公司的细菌dna小量提取试剂盒提取基因组总dna,然后进行pcr验证。
5、dnaassembler获取cm生物合成基因簇
saccharomycescerevisiaevl6-48在固体ypad培养基上进行活化,30℃培养2d,挑取单克隆,接种至3ml液体ypad培养基中,30℃,250rpm,振荡培养过夜。次日转接至50ml液体ypad培养基中,调节od600接近于0.2,30℃,250rpm,振荡至od600接近于0.8,离心收集下层酵母菌体(4℃,4,000×g,10min),用50ml预冷的ddh2o洗涤一次,再次离心收集菌体,加1ml预冷的ddh2o重悬菌体,而后转移至已灭菌的ep管中,离心收集菌体(4℃,4,000×g,1min),加1ml预冷的1m山梨醇洗涤菌体,重复3次,最后将菌体重悬于200μl的1m山梨醇中,50μl/管进行分装,而后准备进行电转化。
准备用于电转化的dna片段:捕捉载体pcap-cm-lr先用ecori进行线性化处理,回收后用ciap进行5’端脱磷处理,最后得到线性dna片段;其他四个片段通过pcr获得高质量的dna片段;相邻两个片段间的重叠区为100~800bp;将500-600ng的每个片段(大小约4k)和2μg的线性载体片段混匀,加乙醇进行沉淀,最后溶于4μlddh2o。
将4μldna片段与50μl酵母感受态细胞混匀,加入预冷的电击杯中进行电转化(电压:1.5kv),而后立即加入1ml30℃预热的ypad液体培养基,30℃,250rpm,振荡培养约1h。离心(17,000×g,30s)弃上清,加1ml室温的1m山梨醇洗涤菌体重复3次,最后将菌体重悬于1ml1m山梨醇中。分别取100μl、300μl和600μl菌液均匀涂布在sd-trp平板,在30℃培养箱中倒置培养约2~3d后,可见克隆子产生。
挑取单克隆在sd-trp平板上划方格,30℃培养2d,进行菌落pcr验证。验证正确的单克隆接种于3mlsd-trp液体培养基中,30℃,250rpm,振荡培养约18h,离心收集菌体后,加入蜗牛酶(2u/μl)10μl,37℃温育30min~1h,离心收集原生质体,而后提取质粒转化e.colidh5α,挑选单克隆,提取质粒进行pcr和酶切验证。
实施例1、创新霉素合成基因簇的生物信息学分析
我们首先对cm产生菌济南游动放线菌(actinoplanestsinanensiscpcc200056)进行dna提取,利用全新三代测序pacbiorsii平台结合二代测序illuminahiseq4000平台对其全基因组dna进行测序,拼接组装获得其基因组精细图(该高通量测序工作由北京华大基因(beijinggenomicsinstitute)完成)。a.tsinanensiscpcc200056的基因组包含一个大小为7,685,618bp的线性基因组和一个13,534bp的环状质粒,g+c含量分别为70.3%和69.0%。经基因注释分析后发现,基因组分析含有7,060个编码基因,其中7,041个位于染色体上、19个位于环状质粒上。
对cm产生菌a.tsinanensiscpcc200056基因组进行蛋白序列同源分析发现,在全基因组中存在唯一的thig(噻唑合成酶)同源蛋白(ats3059)、唯一的moez同源蛋白(ats6133,scp活化蛋白)和几个scp同源蛋白(ats3085、ats3815、ats4181、ats4502、ats4708),而其中ats4181与色氨酸trna合成酶(trprs,吲哚霉素生物合成基因簇中ind0亦为trprs)的同源基因(ats4186)相邻,在此附近还有吲哚霉素生物合成基因簇中存在的色氨酸氨基转移酶(ind8)的同源基因(ats4179),因此初步将cm的生物合成基因簇定位于此,命名为cxn基因簇(图2)。分析cxn中的序列:cxna编码依赖于维生素b12的自由基s-腺苷甲硫氨酸家族的c-甲基转移酶;cxnb编码色氨酸氨基转移酶,与indolmycin生物合成中的ind8[11]及thienodolin生物合成中的thnj[14]同源;cxnc与pane、apba同源,编码2-脱氢泛解酸-2-还原酶;cxnd编码细胞色素p450类的氧化还原酶;cxne编码scp;cxnf编码未知功能蛋白,与thienodolin生物合成中的thnf[15]同源,分析三维结构时发现其含有mpn结构域,属于jamm(jab1/mpn/mov34)金属蛋白酶家族,并且含有保守的jamm基序(jammmotif,exnhs/thx7sx2d,x代表任意氨基酸);cxnt编码转运蛋白;cxnr编码lysr家族转录调控因子;trprs编码色氨酸trna合成酶(表3)。
表3、创新霉素生物合成基因簇及功能
ats4177功能分析:
ats4177通过blastp未找到已知功能的同源蛋白,但通过在线的hhpred对三维结构相似性进行分析时发现与分子伴侣pqqd具有很高的同源性,pqqd可以与pqqe(编码自由基s-腺苷甲硫氨酸家族的酶)相互作用从而参与或影响其功能,因此推测ats4177可能通过与cxna(为依赖于维生素b12的自由基s-腺苷甲硫氨酸家族的c-甲基转移酶)相互作用参与甲基化过程。
ats4183(cxnf)功能分析:
通过蛋白序列比对并未发现与之序列相似的已知功能蛋白,但分析此蛋白的三维结构时发现其含有mpn结构域,属于jamm(jab1/mpn/mov34)金属蛋白酶家族。jamm蛋白可以分为两类:一类为jamm/mpn+亚家族,是金属蛋白酶,具有催化活性,并且在活性中心含有保守的jamm基序(jammmotif,exnhs/thx7sx2d,x代表任意氨基酸);另一类为jamm/mpn-亚家族,缺少催化活性,只作为多亚基复合物的一个组分。有报道发现有一些jamm蛋白酶参与硫转运途径,比如萤光假单胞菌(pseudomonasfluorescens)中合成含硫的thioquinolobactin(一种铁载体,siderophore)需要qbse(scp)的参与,但qbse蛋白在其c-端的双甘氨酸基序(gg,diglycinemotif)后还有两个氨基酸残基(cf),即以前体形式存在,而jamm蛋白酶qbsd可以将qbse前体蛋白中的这两个氨基酸残基水解掉以利于scp激活蛋白(moez)进行后续的腺苷酰化,实现硫转运功能。将cxnf与已知的jamm/mpn+亚家族蛋白(包括qbsd)进行序列比对发现cxnf中也含有保守的jamm基序,而在cxn基因簇中scp(cxne)的c末端在双甘氨酸基序(diglycinemotif)后还有10个氨基酸残基,因此我们推测cxnf的功能是水解cxne的c末端的10个氨基酸残基,释放双甘氨酸基序以利于进行腺苷酰化,从而激活cxne。
实施例2、调节基因cxnr的功能分析
在cm生物合成基因簇内存在一个可能的途径特异性调节基因cxnr,基因全长942bp,编码313个氨基酸。将cxnr的蛋白序列在genbank中进行blastp分析,发现与amycolatopsisluridanrrl2430中的lysr家族转录调控因子同源,一致性为37%;与streptomyceshygroscopicusvar.ascomyceticusatcc14891中安莎霉素生物合成的正调控蛋白fkbr1具有35%的一致性[16]。在产生菌对此调节基因进行过表达,分析创新霉素的产量,初步判断此基因簇是否负责创新霉素的生物合成。
载体pl646[17]来源于整合型质粒pset152,其不含链霉菌复制子,含有φc31attp位点,可以整合至链霉菌基因组中的attb位点,在多克隆位点上游含有强启动子erme*p和tuf1基因的sd序列。克隆cxnr的编码区至pl646的ndei和xbai位点,经酶切鉴定正确后得到重组质粒pl-cxnr(图3)。
cm的产生菌济南游动放线菌属于稀有放线菌,能形成孢囊,孢囊孢子微游动,在isp2固体培养基上于28℃培养7d时,孢子在孢囊内,故而不适合采用传统的接合转移方法进行遗传操作。经调研和实验,成功建立了a.tsinanensiscpcc200056的遗传操作系统(见材料与方法部分)。将过表达质粒pl-cxnr和对照质粒pset152通过转化导入甲基化缺失的e.coliet12567/puz8002菌株中,然后通过菌丝体接合转移导入a.tsinanensiscpcc200056中,从而获得过表达菌株200056/pl-cxnr和对照菌株200056/pset152。提取重组菌株的基因组dna,进行pcr验证。验证引物(pset152与attb_streptomyces)一端在基因组上,一端在质粒上,预计大小约1.6kb,如图4所示。
将验证正确的过表达菌株200056/pl-cxnr、对照菌株200056/pset152和野生型菌株cpcc200056利用isp2培养基进行固体发酵,将新鲜孢子悬液作为种子涂布接种于isp2平板(直径9cm,含25ml培养基),每块isp2平板的接种量一致,约5×105个孢子,而后在28℃恒温摇床间中倒置培养7d,然后将每个isp2平板培养物切碎成小方块,用2倍体积乙酸乙酯萃取48h,最后将乙酸乙酯萃取液浓缩至相同体积(250~500μl),获得固体发酵产物。首先对各发酵产物进行tlc检测分析(图5),结果发现对照菌株200056/pset152与野生型菌株cpcc200056中的cm条带灰度相当,而过表达菌株200056/pl-cxnr中的cm条带则明显加深,提示cm产量有所提高。
为了进一步定量分析各菌株发酵产物中的cm产量,我们进行了hplc分析。利用峰面积进行相对定量分析(图6),结果表明200056/pl-cxnr中的cm产量比200056/pset152中的cm产量提高了98%,此结果表明cxnr参与了cm生物合成的调控,可能为其途径特异性正调控基因,提示其所在的基因簇是cm生物合成基因簇。
实施例3、cm生物合成基因簇及cxnd基因的功能分析
为了进一步确证cm的生物合成基因簇,我们采用近年来发展起来的克隆大片段的新兴技术─dnaassembler[18]克隆分析的cm生物合成基因簇,然后导入s.coelicolorm1146[19]中进行异源表达分析。dnaassembler是利用酵母菌中高效的同源重组机制来一步获得化合物的生物合成基因簇。本工作中将基因簇左右两端约2kb片段作为捕捉臂插入载体pcap01[20]中即得到pcap-cm-lr,其中pcap01包含酵母菌元件(arsh4/cen6复制子和trp1筛选标记)、大肠杆菌元件(pucori复制子)和链霉菌元件(φc31整合酶及其整合位点attp、dna接合转移起始位点orit)使其可在三种菌之间穿梭。将含有cm生物合成基因簇(ats4175-4190)的序列分成4个片段设计引物,通过高保真dna聚合酶进行pcr得到dna片段后,与线性化的捕捉载体(pcap-cm-lr)一同通过电转化导入酿酒酵母中,利用同源重组进行拼接(相邻两片段之间均有一定的重叠),得到含有cm生物合成基因簇的重组质粒pcap01-cm(图7a)。
从酿酒酵母中提取pcap01-cm导入大肠杆菌中,经pcr和酶切鉴定正确后(图7b、c),利用pcr-targeting[21]将卡那霉素抗性基因替换为阿普霉素抗性基因,然后将质粒进行测序,结果发现cxnd(编码404个氨基酸)中由于pcr引入了一个点突变,造成第149位氨基酸由原来的glu(gaa)突变为终止密码子(taa),因此将该质粒命名为pcap-cm(δcxnd),而后通过接合转移导入s.coelicolorm1146[19]中得到异源表达菌株m1146/pcap-cm(δcxnd),同时将m1146/pset152作为对照。
考虑到cxnd基因中引入了点突变,会由于提前终止蛋白表达而造成功能失活,因此将完整的cxnd基因导入m1146/pcap-cm(δcxnd)中进行功能回补。由于pcap-cm(δcxnd)是整合在基因组的φc31整合位点上,因此将cxnd的编码区克隆至含有φbt1整合酶及其整合位点的pij10500[22]载体上,同时在cxnd的上游引入组成型强启动子erme*p即得到重组质粒pij-cxnd,测序正确后通过接合转移导入m1146/pcap-cm(δcxnd)中获得含有完整的cm生物合成基因簇的异源表达菌株m1146/pcap-cm,同时也将pij10500导入m1146/pcap-cm(δcxnd)中获得对照菌株m1146/pcap-cm(δcxnd)/pij10500。
质粒pcap-cm(δcxnd)中仅在cxn基因簇cxna~f中存在4个ecori酶切位点,用ecori酶切后的大片段自连产生的质粒大小约23kb,删除了大部分的生物合成基因(包括cxnabcdef),约5.1kb,将此质粒命名为pcap-cm(δcxna~f)。经pcr和酶切验证正确后将其通过接合转移导入s.coelicolorm1146中得到阻断菌株m1146/pcap-cm(δcxna~f)。所有重组菌株均提取基因组,而后经pcr鉴定正确(图8)。
将所有重组菌株m1146/pcap-cm、m1146/pcap-cm(δcxna~f)和m1146/pcap-cm(δcxnd)、对照菌株m1146/pset152和野生型菌株在isp2培养基中同步地进行固体发酵,经乙酸乙酯萃取得到发酵产物,然后进行lc-ms分析。
发酵结果分析,
一、cm生物合成基因簇功能分析
与对照菌株m1146/pset152相比,m1146/pcap-cm的hplc峰形图上存在明显差异(图9a-ii、iii),在18.1min和16.9min处都出现了差异峰(1、2),提取分子离子图发现[m-h]-分别为232和218(图9b),与野生型菌株cpcc200056中的cm与dcm的出峰时间和分子量相同(图9a-i),并且两者的uv最大吸收峰为230nm和300nm(图9c)。而在阻断菌株m1146/pcap-cm(δcxna~f)中则没有出现这两个峰(图9a-iv)。以上结果说明cm生物合成基因簇在异源宿主s.coelicolorm1146中成功表达产生cm及其类似物,确证其为cm生物合成基因簇。
二、cxnd基因功能的分析
在cxnd基因功能缺失的异源表达菌株m1146/pcap-cm(δcxnd)及m1146/pcap-cm(δcxnd)/pij10500中,没有发现产物cm和/或dcm,但是在18.8min处出现一个新峰(3),出峰时间与cm接近,其分子离子峰m/z234[m-h]-,比cm(m/z232[m-h]-)多2da(图9b)。而且,此新化合物的uv图谱显示其峰形与cm相似(图9c),最大吸收峰为220nm和280nm(cm的最大吸收峰为230nm和300nm),其中波长较大的特征峰为cm中富电子基团(如o,s等)的芳香大共轭系统π-π*跃迁引起,若失去与芳香环相连的富电子基团,该特征峰将发生蓝移(向短波方向移动)。结合化合物cm化学结构,我们推测新出现的化合物可能是在生物合成过程中cm的芳香环外c-s键没有形成的产物。这一推测得到了lc-hrms/ms结果的确证。
为了解析新化合物的结构,我们将此差异峰进行收集浓缩,而后利用高分辨质谱进一步分析,在负离子模式ms/ms图中(图10b),分子离子峰为m/z234.0600,对应分子式c12h11no2s(理论值:234.0606[m-h]-),经过一定能量裂解之后得到了多个碎片离子,其中丰度较大的三个碎片离子分别为115.9555、141.9682、173.9387,根据cm化合物同等条件下的ms/ms结果可以推导出该化合物裂解过程(图10),因此该化合物的结构如图所示(图10b与图1),为cm的开环结构(secochuangxinmycin,scm,图1)。该结果提示cxnd基因编码的cxnd蛋白参与c-s键的形成。
实施例4、创新霉素生物合成过程分析
结合上述实施例1-3的结果和生物信息学分析结果,对cm生物合成过程做如下分析(图11):
图11硫载体蛋白的活化(a)和cm生物合成途径分析(b)。
1、jamm金属蛋白酶家族的cxnf水解cxne(scp)末端的10个氨基酸残基后露出双甘氨酸基序,而后先在scp活化蛋白moez(n-端为类似于scp活化蛋白thif的结构域)作用下生成腺苷酰化的scp,再在moez的c-端硫氰酸酶结构域的作用下形成硫醇化的scp,即为硫原子的供体。
2、另一方面,前体色氨酸(l-trp)中的氨基在氨基转移酶cxnb的作用转变为酮基形成吲哚丙酮酸,然后在c-甲基转移酶cxna的催化下形成3位甲基化的吲哚丙酮酸(3-甲基吲哚丙酮酸),再在还原酶cxnc的作用下将酮基还原为羟基形成3-吲哚-2-羟基丁酸,而后在噻唑合成酶thig的作用下将硫醇化scp中的巯基替换羟基得到3-吲哚-2-巯基丁酸,即开环cm(scm),最后在p450氧化还原酶cxnd的作用下形成c-s键,得到创新霉素。
最后需要说明的是,以上实施例仅用作帮助本领域技术人员理解本发明的实质,并不用做对本发明保护范围的限定。
【参考文献】
1.chuangxinmycinresearchgroup:studiesonanewantibiotic-chuangxinmycin.scientiasinica1977,xx:106-112.
2.guzp,liangxt:thestereochemistryofchuangxinmycinactachimicasin1985:250-256.
3.guoxl,zhangzp:anewtotalsynthesisofchuangxinmycinandthestudyofitsstereoisomers.yaoxuexuebao1987,22:671-678.
4.sush,tujd,zhangsw:synthesisofsomederivativesofchuangxinmycin.chinjpharm1984:17-21.
5.wangyc,xuxd,zhangzp:studiesonnewantitumoractivitiesofchuangxinmycinderivatives.chinjantibiot1992:417-421.
6.qitq,liux,yandyf:repressiononthebiosynthesisofenzymesinvolvedintryptophansyntheticpathwaybychuangxinmycinzhongguoyixuekexueyuanxuebao1980,2:32-37.
7.caoj,qitq:astudyonindolepyruvicacidmethyltransferaseinchuangxinmycin-producingstrain.weishengwuxuebao1989,29:63-67.
8.zuolj,zhaow,jiangzb,jiangby,lisf,liuhy,yuly,hongb,huxx,youxf,wulz:identificationof3-demethylchuangxinmycinfromactinoplanestsinanensiscpcc200056.yaoxuexuebao2016,51:105-109.
9.xuj,may,liy:studiesonthebiogenesisofsulfurinchuangxinmycinmolecule.actamicrobiolsin1978:66-70.
10.zhouxz,linl:vitaminb12playsanimportantroleinbiosynthesisofchuangxinmycin.zhongguoyixuekexueyuanxuebao1984,6:109-111.
11.duyl,alkhalaflm,ryanks:invitroreconstitutionofindolmycinbiosynthesisrevealsthemolecularbasisofoxazolinoneassembly.procnatlacadsciusa2015,112:2717-2722.
12.sasakie,zhangx,sunhg,lumy,liutl,oua,lijy,chenyh,ealickse,liuhw:co-optingsulphur-carrierproteinsfromprimarymetabolicpathwaysfor2-thiosugarbiosynthesis.nature2014,510:427-431.
13.parkjh,dorresteinpc,zhaih,kinslandc,mclaffertyfw,begleytp:biosynthesisofthethiazolemoietyofthiaminpyrophosphate(vitaminb1).biochemistry2003,42:12430-12438.
14.milbredtd,patalloep,vanpeekh:characterizationoftheaminotransferasethdnfromthienodolinbiosynthesisinstreptomycesalbogriseolus.chembiochem2016,17:1859-1864.
15.wangy,wangj,yus,wangf,mah,yuec,lium,dengz,huangy,qux:identifyingtheminimalenzymesforunusualcarbon-sulfurbondformationinthienodolinbiosynthesis.chembiochem2016,17:799-803.
16.songk,weil,liuj,wangj,qih,wenj:engineeringofthelysrfamilytranscriptionalregulatorfkbr1anditstargetgenetoimproveascomycinproduction.applmicrobiolbiotechnol2017,101:4581-4592.
17.hongb,phornphisutthimass,tilleye,baumbergs,mcdowallkj:streptomycinproductionbystreptomycesgriseuscanbemodulatedbyamechanismnotassociatedwithchangeintheadpacomponentofthea-factorcascade.biotechnollett2007,29:57-64.
18.shaoz,luoy,zhaoh:dnaassemblermethodforconstructionofzeaxanthin-producingstrainsofsaccharomycescerevisiae.methodsmolbiol2012,898:251-262.
19.gomez-escribanojp,bibbmj:engineeringstreptomycescoelicolorforheterologousexpressionofsecondarymetabolitegeneclusters.microbbiotechnol2011,4:207-215.
20.yamanakak,reynoldska,kerstenrd,ryanks,gonzalezdj,nizetv,dorresteinpc,moorebs:directcloningandrefactoringofasilentlipopeptidebiosyntheticgeneclusteryieldstheantibiotictaromycina.procnatlacadsciusa2014,111:1957-1962.
21.gustb,challisgl,fowlerk,kiesert,chaterkf:pcr-targetedstreptomycesgenereplacementidentifiesaproteindomainneededforbiosynthesisofthesesquiterpenesoilodorgeosmin.procnatlacadsciusa2003,100:1541-1546.
22.dud,wangl,tiany,liuh,tanh,niug:genomeengineeringanddirectcloningofantibioticgeneclustersviaphagevarφbt1integrase-mediatedsite-specificrecombinationinstreptomyces.scirep2015,5:8740.
23.heinzelmanne,bergers,puko,reichensteinb,wohllebenw,schwartzd:aglutamatemutaseisinvolvedinthebiosynthesisofthelipopeptideantibioticfriulimicininactinoplanesfriuliensis.antimicrobagentschemother2003,47:447-457.
24.kiesert,bibbmj,buttnermj,chaterkf,hopwoodda:practicalstreptomycesgenetics.norwich,england:johninnesfoundation;2000.
25.kornf,weingartnerb,kutznerhj:astudyoftwentyactinophages:morphology,serologicalrelationshipandhostrange.ingeneticsoftheactinomycetales.freerksene,tarnoki,thuminh(ed).newyork:gustavfisherverlag;1978.
26.sambrookj,russelldw:molecularcloning:alaboratorymanual,3rdedn.coldspringharbor:coldspringharborlaboratory2001.
27.pagetms,chamberlinl,atriha,fostersj,buttnermj:evidencethattheextracytoplasmicfunctionsigmafactorσeisrequiredfornormalcellwallstructureinstreptomycescoelicolora3(2).jbacteriol1999,181:204-211.
sequencelisting
<110>中国医学科学院医药生物技术研究所
<120>一组生物合成创新霉素或开环创新霉素的基因簇
<160>39
<170>patentinversion3.3
<210>1
<211>21665
<212>dna
<213>actinoplanessp.
<400>1
tcagcttgtcgtggcggtggcgccggcggttccgccgctgaccagcttggcccaggtggc60
ggggccggcgatgccgtcggaggtcaggcccttggccttctggaaggcggtgagcttgct120
ggtggtggccgggccgaagacgccgtcggcggtgacgtcgtagccgttgtcggcgagctg180
gcgctggagggcggtgacgtcggtgcccttcgagcccgacttcaccgtggcgatcagctt240
cgcccaggtggcgggaccgaccatgccgtcggcggcaaggccctcggccttctggaacgc300
ctggaccttcgcggctgtccccgtaccgaacacgccgtcagcagtcgtcgcgtacccgtg360
cgcaccgagcagcaactgcacggtcgccacgtcgacgcccttgtcgcccgccttcaccgt420
cggccacgacgtgcccggctgcggctgcggcgcaccgcctcccttggcgagggcccgcag480
ctggtccaggctgccccggtacacgttgcggtcgcccttgcccgcggcgccgttcttgcc540
ggggaggcctgggatcgcctcggactccgtgtactgccagagcgaccaggcacccgcgcc600
cggcacgtcctgcggctccttcgacccgctttcgtagcgggccagccacagcgggtggtc660
cttgaagacctggcccttgccggccatgcagccgttcacgaacgacgcccgtgtgtagac720
gatcggcgtcaccttgaacgcctcctccacgcggttcaggaaggcggtgagctggtcggc780
gcggagcgccttcgggcacacctccttgccgttcacccacgtgccctcgacgtccagaac840
cggcggcagctccccggccctcttgccggtgtagccggcggaccgggccgcgcggatgaa900
gtggtcggcctgcgcgccgccgtccgtggtgctcttcgggtcgaagaagtggtacggggc960
gcgcagcagcgatgtgccggacgcgtccttgaagtcccgtgcgaaccaagggtccttgta1020
accggtgccctgcgtcgccttgaggaacgcgaaggagttggactgggcgacgcgcttcca1080
gtcgatgggcttgccggtcgcgtcgtggttgtggtggctggtgtcgacgcccttgacctc1140
gtacgtactgggaggggcggccacgctctcgtccgcggtggacagcaggacaccactcat1200
gagcgacgcggtggcgacggccgccgcggccaggcgcagtccgtgacgacggccgcgcgg1260
gttctccatggacacagtcaggtcccctttctgaggcatgtgggcgcaaacccgcgcacg1320
ccgcagcgggcatgacggacgggttcaagtggtggtcgaacgaccggaattgggggtggt1380
tgccgtaaccatgcaccggcgtcaccagcgagtcctacactttcggctggccggctgatt1440
catgcacaaggcccgggtgatgcagggcggttgagctgccgtcatgcacggtgaacttgg1500
ggggacgcataaatgggcgttgaacgcgacgagccgacgcggtcagcgaggcgggaactc1560
gccctgctgttgcggagctggtgggaggcgcaccccgacaagatcacgcaggaggcgctg1620
gcccggcggatcacggagcggggcgtacggatcagccaggagatgctgtcgcgctacctg1680
aaccggtcccgcccgaccacggcccggcccgacgtgatccgcaccatgcacgaggtgctg1740
cgccgggcgccggaagagctggacgtggccctggaactgcacgctcgggccaccgccccg1800
cagacgccgcccgccgagggggcggccacgagccagccggccggggacgcggggaccgcc1860
gcgccgaagggcgtagagccgacctcggcggccccgctcttgacccgcacgccccacacg1920
ccccgccccgcttcgcggaagaagtggccgtggatcgccgtcgtcgcggccgcggtcgtc1980
ggcgcgtccgggctcaccgccttcatgacactgggcgaccagcggcagaacaccccgcgg2040
ggacacggagcgacaccctccgcctcacccaccgccctggtgtcacccaccgcccagggg2100
tcgcccgccggcacgcatcctcccgcggagtgccgcgacgagtcctgcttcggcatcgac2160
gccaagtacgccatctgccaggacgacgccgccacttactacacgggccgcgcccacggc2220
gtcctcgtcgagctgcggttcagccccgcctgccaggcggcttgggccaagatgagcggc2280
acctcgcagggcgatgtcgtacgcgtcaccaacaacgcgggccgcagccgccactacacc2340
cagcagtggggccgcgacgcccacaccacgatggtggaggccgtgagccccgacgacgcc2400
aaggcttgcgcccgcaccccgcgcggcgaggtgtgcgccacgaaggccgtcgcgtccgcc2460
ccgcgcgacgcggcacctggcgagcgcgcggcacctggcgggcgctgacggccccggatt2520
cctggccgccggccggacggtgtcgccccagggcccggggacgccctacataaccgcagt2580
tcagcggcgtgcatagcgggaagctcatgcatgcatcaatgcaccgccgtagcgctggat2640
cacgtcgcctccagccgtgcagagtagagccgcggccgacgagcccgctctctccctcct2700
gggatcgaggtgcgggtggatccgcgatcccgagcgtgccgacgtacgccggtaccggcc2760
gccgcccctgcgccacacgcggacgcagggcgcagaccactgccacccaaggccaaggag2820
tcagccatgcccgcacgcaccacacgaaccgcacacaccacacgcaccggccggttggcc2880
gtcgtcgccctcgcggccttgacctgtgcgggcctggtcaccggaactgcagccacggcc2940
accacacccgactccctgcccaccgcgaagcgcgccgcagcgcccgacgcagcggctgta3000
tcgtggccgacgctgaaggcgggcgcgcgcggtacggaggtgaccgcgctccagcacctg3060
ctgatcgcccgcggccaatccgtcgccgtggacggggagttcggcccggccaccaccacg3120
gccgtcaaggcgttccagaaggccgacgggctcaccgccgacggcatcgtcggacccgcc3180
acctgggccaagctcgtcccgacgctgcgtcagggcgcgcagggcgcggcggtgaaggcg3240
gcccagaccctgctgaagacccgtggccaatccgtcgccgtggacggggagttcggttcg3300
gccaccacctcagccgtcaaggcgttccagaaggccaaggggctcagcgccgacggtgtt3360
gtcggcacgcagagctggtccgcgctcctcacctcggactccggcgcgccgtccgggaac3420
cgggccgcgttcgcccagcagatcctcaacaccagcggcatcgagctggcgaccgtccac3480
cccggcggcacccacgccggctccaccgcccggcagaacatcatcgacacagccaacggc3540
aagggcgctctgaccagtccctggagcgacaagccgaaccagcgcgtggcgctcgacacc3600
cggatgctcaacgggctgctgaagctgctctcccaggacggctaccggatctctgtctcc3660
gagatcgtcggcggcgaccacagcacgaactcccggcactacgcgggactcggcttcgac3720
atcaactacatcaacggccggcacgtcggcgagagcgccccgcaccagggcttgatggcc3780
gcgtgccggaagctcggggccaccgaggtgctcggtccgggcgacgccggccacagccgc3840
cacgtccactgcggctggccgcgctgatcccggctgaccgccaacttcccgtgcctgcaa3900
gcagagagggtccgtcaggaagcatgacggaccccctcatgcaggatcgagggtgacgtc3960
cgggctactccgtagcaccgtccgatttcttgccggctgcggccaaggttcctcccaata4020
gggccgcgcccaggggccatgaacgcatgtgcacggggagggtcacgcagtgatccccga4080
cgcgcatcacaaggcccttgttcaccaaatcgcgtgcggcggcggcgaattgcgagtttc4140
gggtcacctcggcccacgcgcatccctccaggcccaggaggaagacggccatctgggtgg4200
gatcgtccacgatgatttcgcggctggattcggggcgctgatcgacgacggacaggaact4260
tcgggcccttacggaagtagaggaggccgaaattgctggaggaccgccactcaccgacgg4320
acggatgccctgtctccagcaccgtgatgctgtcgggggcaggaaggtgatcgagacggg4380
ggaccaggtcgagctgttcggcgcccagggtcagtgaccaggtgactctggtgccgatcg4440
aggaacattcgcgaatcaacgcgatgatccgcacgatcacgtgactggggagtgccgaga4500
agtcggcgggctccggcagccgtaccccggcgacgggcacccggtgcagtagcgccgcca4560
gttcggtggcgacgggaagttcggcccggtcgagtccgagggtcagcgtctcggtcgcgg4620
gacggccaccggccggtacccgtccctcgtccgaatctcgggccttgagttcatcgatgt4680
ccagcaaagcgctcatagcgaagccgccacctccttgccgccgaccagctttcggcgata4740
cgggtcaaccccgagggcgacgcttacgtagcgcccctcgtcctcgaatgccaggccgcg4800
atcgacgaagtagcggagcatttcctcgagttccgcttccccgacgacgtgcccgctgtc4860
ggcaagccgccggcgtatgccctcgcgggcggcgcactggaacatgccgaggtacacatt4920
gctgcggacctcgtccagctcgatcacttccgtcggccagctggcacggcggtcttcgat4980
gacgacccggcctcggtcatcggtccagtaggaaagggtgccctgcggataggccttggc5040
ccattcctcgcaggcctgcttcatctcgtcctcgatgggccctgagattccccggacgct5100
ggtgtcgaagaagaacaccatgtcgtacagctgatcctgcgggatctggtagatgaagtc5160
gtatatttccgaggggcggcggaacatgaacccctgggtggggtcctcgaagtagggact5220
gaaccgctcaagggctatgcgccaagccccggttggcggctccaggtgctcgagcgtggc5280
caatttcttgagcagcccgcggtagtcgtcctcggtctcgcccgggaagccgtagaggat5340
gctccatgtcacgttgagcccgagatcctgtccgtcacgcagcatccgtacgttgtgcgc5400
ggcactgacgcccttgtccatgaggcgcagcacatggctgctcaggctctcgataccggg5460
ctgcacgaagaggacgttcgcctctttcagcctactcaactgctcccggttcatattgga5520
cttgatctcgtagtgaattcgcagatcgcagtcgagggcagctatctcgggcatggccgt5580
attgagatacttcatgtcgaggatgttgtccaccatgaccaggtcgaggatctggtgtcg5640
ctcggccagttcccggacttcctgggcgatgcgctcaggggccttgctccggaagtcgat5700
attcgatccgttcaggccgcagaacgtgcattggtgagcctctccccaccagcaaccacg5760
ggaggtctcaaggaccagcatcggacggacgtggtgacggacgggtgacctttcgagggc5820
ctgaaagtagctgtcgtaaccgggcgcgggcaccatggcgaacggcagcgccgccgtggc5880
cggtggattcaccaccggatgcccgtcatcccccctccagctgagccccggcacgtcggc5940
gaggctctcgccccggatgatgcgattcagcaacgcgggcagcgcacgttcgccctcacc6000
gctgatcacgaagtcgagttgctcgaaattccggtgcaacgcgggaccttgtgctccgtc6060
gcagttgctgccgccaaggaccgtgcggatgcccggcgcgagtttcttcagctccctggc6120
cagtgcgagcgacgggacgttctgcatgaaggtgctcgtgaacccgaccacgtcgggagg6180
atcggcagcgatctcggccgcgagatcccggatgaatccccgggcgtacttgtgcatctc6240
aacgggaagtgtcgggtccatgtcccgctgctcgaggaacttcgcgtactcgtcgacctg6300
ataactgtcgacgtcgtacagcgctggggtgaacacccagtcccctacgccgtggaagac6360
ttgatccgcgatgttcccgtagtcctcgcaggtgacggagccgttgctctcccgcatcag6420
gtattcggcccagcggaggttggcgtacagctcatcgacggtccagtcggcggcgttctt6480
gcggacgcatggccccagtacgcccagcgcgctggacggcgtgtcgagcccttgccacgg6540
catggcgatcatcaggagtttcacagacttcccaccccagagttgctgtcgatgagttgt6600
atgaggacgggcaaaagaattgtccgcagagagttctcagaaccagttcccacggtccgc6660
ccggccgagcgccgccaacagcgtccgatgtgcctcggcctctcccactgagatccgtac6720
accgtgccccggaaaggctcggaccttgacccctgcggtagccgcagtccgggcgaaaga6780
ctcggcggccgaagcgagcgggagccagacgaagttggctcgggaaagcaggacgggcag6840
cctcagttccctgagttccgcggtcagttcttcgcgtgccgcagccactgctgccagacg6900
ttcacacagttcgtcctcgctgcgcagcgagagcattgcggcttgttccgcgaagcgcgt6960
cactccgaaagggattgccgtcttgcggacggtggccatgacctgccgtggcccggccgc7020
gtaaccgacccgtaggccggcaaggccataggccttggagaacgttcgaagtaccacggt7080
gttgctgtgctcgctcagcaacaccggcagacccggaggattggcgccccggtcgaactc7140
cacgtacgcctcgtcgaggaccgcgaccacatgagccggcagcgaacgcaggaaaccgtg7200
cagctcgtcttggtcaatcacggttccggtcggattgtgcggggagcacaggatcaccac7260
cctggtccgcgcattcacccgggtgcggatctcatcgagatcgtggccgccggacgcagt7320
caggggcacgtggactccggtggcacctgaaatggcgaccaacagcggataggcatcgaa7380
tcccggccagccatggacgacttcgtcgcccttgccgcacagtgcgagaaggatctgctg7440
gagcacgcccgcgcttccggggccgaccgcgacctcatccggggagacgcacaagtgccc7500
ggcaatgtcctcggtcaggtcccgtgctgtggggtcggggtaacgagcaagtcgcggcaa7560
gcctttttcgataccggcaagcacggtaggcagcggggggagaaccagctcgttgctgga7620
caggtcgaaggtgaaccgcgagctgccttcggcgttcgacgactccttgtcccggtaggc7680
ccgcatgtcccgcagggtgctgcgttctgcgaatctcacgttcacgcctgcagactcctc7740
gccgtgcgtgattcgcgggtggcgagcagctcgtagagcatttcgtggaccggcgcggtc7800
aggcccgccccgcgggcgaggcggaccacggccccggtccacgcttcgagctctgacggc7860
cgtcccgccaggatgtcccgttgcagcgaggaggtgacgtcgggcgactgctggtccatg7920
agctctgtcgcggtgtccacggcagctgccggcagcgcgattccgagcttgatcccggtc7980
tcgtagatctcccgcatgccggcgatcagaatgttgcgggtgccggtgcgcgaccggagc8040
tcgccgatggtcgccccgccggtggcggctccaaggctgccgatcgggaccaccaacagg8100
aacttcgcccaaaggccggcccagatgtcgctcggctcgggcacggacaccgaggcagca8160
cgcagcacctcgcgcagtcgtgccacccggtcggacacagtgctgtcccactcggtgaag8220
gccagagcgccggggggacccacgtgcctcaactcgcccggaccggccgtcgaggccacg8280
accctgacgctgccggggagtacccgaccgcggccgatcctggctgcgacctgctcaggg8340
gcttccaccccgttctgcaccgtgaccacggcagtgtgctcgccgaccagcgggcccagc8400
gcgtcgagggccgccggcagctgtgaggtcttgacgcagagcagtacgaagtcgacctcg8460
ccgatgtccttcgggtcggccgacgcccgaacgtccggcacacgtaagtcacttgagccg8520
ttggtgatgcgcagcccctgtcgcctgagcgcggcgaggttctcgccgcgggccaggaac8580
cgcacatcatgcccggcggcggcgagcaggccgccgaaatagccgcctactccgccggct8640
cccaccacagcaatgctcggaccgccttgttccgtcatcgcggcaacctctctcccactc8700
gtatcgggagggcctcgaggccccggctgagcgaactgtcggagagccaggcgacctctt8760
ccggaggcacggcgagctcgaatcgcggcagtcggcgcaacagcgtggagaaggcgatct8820
cgccctggagtcggcccagcggagcgccgatgcagaagtggggaccgtggccgaaggaca8880
gatggcggttcggtgaccgcgtcacgtcgaaaccgtccgggtcgtcgtacacgtcgggat8940
cacgacccgcgctggacagcgacaggtgtacgaagctccccttgggtatgtccactcccg9000
cgagctgcatgtcctccgccgcgacccgcagcgaggcccaggcggccgacccttcgtagc9060
gcaggatctcctcgatcgccgaagggatcagctcgggggtggctcggagcatctcgagct9120
gctggggatggcggagcaacagtgctgtgccgttgccgatcatgttggcgaccgtcttgt9180
gaccggcgatgatcagcaggaggagcgtcgaaaggagctctgtttcgctgtatacgcctg9240
cgtccctggccctgatgatctcgctcagcaggtcgtcccgcaggtccgtgcgccgctcgg9300
ccacgagcttggtgaaatagtccgtgaactcttcgctcgcggctttcagctccgcctcgt9360
cgtgctgcaacgggtcctggctgaggatgtagctccactccagaaacagcggccggtccg9420
ccaccggtatccccaggtattcgcagatgacggtgagcggcatcggcagagcgaaggaac9480
tgagcaggtcgatttcgccgttctccggaaaggtgtcgatgagatcgtcgacgatgtcct9540
ggatgcgcgggcgtagttgggcgaccgtggccggcaggaatgcccggctgatgggcttct9600
tcaggcgggtgtgtttgggcgcgtcggcgaatcccaggttgccgaccacgaggagactgc9660
tggccacggccttgtctcgatagcgggcgggcaggttctcgacttgcttggagaggcgcg9720
agtcgccgagcgcttcctcaacgactttgttcccgaggaccgcgtacgcgtcggcgccgg9780
ggggaacattgatccgatggaccggacacttggacctgtactccgcggctgtcgcgtgcg9840
gattcgaaccgggctcggtgaagaattccgtgggaatcacgtcggtcatcgtgactcctc9900
ggctcgcgcggcttcgctgccacctgctaccgccggcactatccagatgacgtcgtggtg9960
ctccactttcgtttcgaggccgtcgaggctcctgatgtcgctgtcgttccggtagacatt10020
gacgtagcgcttcacggacccttcctggtccatgagtcgctcgaggacccccggacaggt10080
ctggtcgagaccgacaaggacctcccggatattggcgccctcgacaggcaactgccgccg10140
accgccggtcaggacgtggaaggctgcgggaagtttgacatcgggcatttcttattcctc10200
ctcgagtaggacttcgaactgctcatggatcgccgccggaatccgtatgtcaagcctgtc10260
gcattgctcggcaccgggccgccaggccgctacccgatgtgcgataccgccactactctc10320
atgcacgtaccagatgacgttcactggccagcatgactgccggaaaaggtattcatccat10380
ctgtgtcggatgttcgctcaggcgctggcgccgttcgtgcggcgggccgatttcatgcca10440
gttcggatgtgaatgtatggagcccagcaactccaggccgttcgccgactgttgcctgat10500
cgcctgcaagacgccctgttcatcggaccagaatcctcgccccggattcttgtacacgtc10560
gccgaactgcggggcgatcgtcgcttcgaattcggccatgacactctcgtcactgtcccg10620
gacattcggcacgaattccacatcgctgatcactatttcagcgccgcccacctgcccgaa10680
caacagacccgatgcacgcggcagatggctcgggacgccatcggcgtcgcggctgtccag10740
gcatttttggtactcccccagtgcgctgctcaggaaacgcctgacgggttcgatttcaaa10800
tcggacggtcaccggcagatcgtcgagcagggctgcactgtctgctgtgccacccagacc10860
gtcatccgtagcagctgtcacatactgggcatctttcacgcctgccatttacaccctccc10920
tgagatttcctggtgaaatccttaggccctgggcagtcggtggaattgcggagcgttcat10980
gggcgtacgccccctgcgctctggcgaggcaggtcagtcacccctggcatacgagggccg11040
cggagcgtggaggtcttgatgccgacccagcacaacagccctccggccgccgctacggcg11100
gcagcgatccagaaggcggtggtgaagccgtcgcgcgccggcgtatcggaaccggagata11160
tgccaggcagccagcaaggcgacagccagctgactgcctacgacgccgcctgccgtgcgc11220
accaccgtgttgacgccattggcggttgcggtgcgtcgggcctccgtcaagtcgctgatc11280
accgacgggagtccgctcagcaccagtccggaaccgagacctacgaccaggtaaccgacc11340
gccacttgccacccgttggcattccacgcccagagcgagatcgccccgaccgccatcacg11400
gcgaagcctgatgcgagcgtcgcgcgtacggacgtcaggcgctgcagcagacctgctagt11460
ggtcccgcaggcaacagcaccaatgagccgggcagaaggagcagacccgccatggtgaca11520
tcggccccgagcccgtagccgatggtccctccgcccggtagccgctgatccgccgcggtc11580
tgggcgtacgtcggcagcaggacgtagaacacgaacgagaccacgccgaacacgaacgcc11640
gcaccgtgcaccgagacgaaggagcgaccggccacgaccgcggggtcgatcaagggggcg11700
ggcgactttcgttcgactacgaccagcaatcccaggagaaccgcggaggccccgaacaga11760
gccagcgttcctgtggacgcccacccccatgaggtgcccttggtcaacgcgagcaacagc11820
gcgacgaggaccaaggccaggagcaccgccccgggcacgtccaccggttcgccggcttcg11880
ccccgctggtccggaacgtacttcgcaacgagcccgatcgcgcccaggatcagtaccgcg11940
gcgacggcgaacagccagcgccatgactggtggtcgacgaccagacctcccacgaccagg12000
ccgatgccggcccccacaccgatggtccctgacaccagcccgaggcccgaacgcagccgc12060
tgctcaggaagtacgtcgcgcaggatgccgaaggacagggggatggccgcgaggctgact12120
ccctgcacggcacggcatgcgatcagaaccccgatattgcctgccacagcacaaccgacc12180
gtgccgatcaggtaggtggtgagaaccagcagcaaaactttgcgcttgctgtagcggtcc12240
cccagccggctcagcaagggcgtgctcgccgcactggtcagcaggaacacactcaggatc12300
cacgccgaccacgtcgaggccgtgtgtaactgcacctgtagtacgtgcaaggccgggacg12360
accatggtctgcatgagtgcgtacgacagcacggagacgccggtggcgatcagcgtcatc12420
gtcgcgctgatcgacggtggacgtggcgaagtgccgttgtaggacaagggggacccccgt12480
ggggatgtggtcggggcggttactgagtcacgagaccactggccggagccacatcactga12540
ggcttaaacactgcctgcgcaaagaatccgtcaattgtgtttggggcgatacacccgctg12600
aaagcgacttcgtacggcaaagacaagaagacctccggtggaattttccacaggccccgc12660
agtcaaggctgagattcgatagctgcaccttgcatgggtgatcgtatgtagccgcctaca12720
tacacgccaatacccgttgcgtatggggccatatactgatcatatggaactggatttgcg12780
gcacctcaggtacttcgttgccgtagccgaggaaggcgggttcacgcgagccgcggcccg12840
cctgcacatgacacagccgccgttgagcgtggcgattcgtcaactcgaaagagagctggg12900
tctccagcttttggacagaacgggcaacagagtcgaactcacgtcggtcgggcgcgactt12960
cctgactcacgcgaggaacttgttgcagcagtggcaggtcacggtcgagaggatgcggca13020
ggcggggtcgcaggatgtcgaacggctcgtcgtcgcgttccgcccggccgtcagccgccc13080
tctggcacaccggaccattgaactcatccgcgaaaagcaccctgagtatcaggtagtgcc13140
ccggtacgtaccgtggaccgaacagacagcatgcctggaggcaggggacgctgacgtgtc13200
cttcgtgctggagcccgcggactacgtgggcctcgagagggccaccgtggccctgttacc13260
ccgggtcgtctgtctgccatcggctcacgagctggccagtcgtgactccgtgtcgatcga13320
cgacctgagcgaggttccgatcattcgccccaccggcgggtcgcccgagtggtccgactt13380
ctggggtggtgaggtgtgccccggcaagcgcacctggaaggaacctcccacagcgacgcg13440
cctcgacgaggccatcgacctcgtggccctcgagaacgcagccgcgctcgtccccgtctc13500
tgtcatggcagtccagcaccgtcaggacgtcgtcttcatccctgtgacggatgtgcctgc13560
cgcccggttgtcccttgcctggcgtgagggttccgactccgaactggtacgcctcgccgt13620
caggtgcgctcaggccgcagcccaggatccagccgtcaggacgctcttcggagaacctcg13680
accaaccggaaccgctccggcctgatgagagcaggggctgtcaggaacttcctgatggcc13740
cctgccagttgtggtacgttcgcgatcatgttgcgcaggaatctttgttggtgggccgcc13800
tagggcggcccaagaacctgcgctcgaccgagccagccagccgctccgagggacgggctg13860
ctgaacttggtcggcccgcctcgtcgatgcggtcaagacgagaggatcagggccaatgtt13920
cagtctttccgggatcacgccttcaggtaaggcgcacctgggtaactaccttggggcagt13980
gcgtcgttgggcagcacagtcgggccccgaagacctgtatttcgtcagcaacctgcacgc14040
catgacgaccaagcacgaccccgaacgtctccaggaactgaccgaccaccaactcgcttt14100
actcatcgcggcgggcgtaccccaggaacgtctcttcgtgcagtcggacctcatccagga14160
gcacatggcgttgacgtggcttctcgagtgcacctgcaccttcggggaggctcgaaggat14220
ggtgcagttcaaggagaagtcccaggggagcaactccgtacgccttggtctgctcaccta14280
ccccgtcctcatggcggcggacatcctgcttcatggcgcttcagaggtgcccgtcggtca14340
cgatcagaaccagcatgtggagctggcccggaccttggcgcggcggttcaacacggacta14400
cggcgaggtgttcacggttccgcaagccgtcctgcccgtagccgcagcccgggtacgtga14460
tctcgctgcccctacgcggaagatgtcgaagtcgtcctcggacggcagcggcatcgtcta14520
cgtcctggacagcccggaggccgtacgccggaagttccaacgcgcagtgacagacggaga14580
aaacaccgtccgctacgccccggacgaacagccgggcgttgccaacctcctggagatcag14640
ggctgcctgcactgacacgctcccgagcgatgcggcgaagggtatcgattcctaccgtga14700
cctcaaggaagcagccgcagaggcagtgatctccctgatcgcaccggtgcgtgagcgggc14760
actgcagctcctcgaagagcgatcggagctggcgaagatccgggctgagggggccgaccg14820
tgctcgggcgcggtcacgagaccgcttggatcgtgcgctcagccttgccggtctgaagta14880
gcagatcaccggccgggctctggctcacaggagctccggagtccggccgggctctcgatc14940
aagtgatgcgcccgagggtccctcatgcgtaaagatgtggcgcaacggatgccagtgggg15000
ggagcagatctgttgcgtacgcgcgggattgccggggtcgtgtcggcggtgctgggcgtt15060
cttctcgcgatatcactcgcaactgcccccgcccatgcggcagttcgctcggccgcggcg15120
gtcgatgtctgtcggtcggccgccctgagcaaggcgcgtgtgagcacgtgggtgcggctt15180
gagcaccgcgatggtacgtacagcaggatccgcagcgagctcagcgtcgaggtgcccgag15240
gattggccgttggccaaggacctgctgctgagtgaggacagccgccggtacgtcgcggcg15300
atgtcctgcctcacccgtaccgatcggggccggcaacgccgctggtcggagtggaggagc15360
agccgtccgacggtggcgtccacgaagagcggtggggtgaaggtcgtcgaccgtacgcac15420
tcctgggtcaacgtgtatcgggcgcacatcgatgtgggtacctggcgggtccgtgcgggt15480
gcggagcgctggaccgtacaactgcaagctccgtccgcgctgaacgcggcccgctgggat15540
gagatcagggtggaacccggcgccccgggagccgagtcggcgaccccgcggcctgacgag15600
gggcgcggcgccacggcgttggtgtggcatccccagaaccaccgtgagaaggcggctgct15660
cctgccgtgagcgttgcgctcaagccctcctggcagcgttcgtgggcagcccagaacgac15720
cggctggtcgccgtggcgctggatcggggcggatggctgctctgggacgcgacgagtgcc15780
gccctgttgctgtacgcaaccgtcctgtaccggaggcgttccgctcctcccactcaggct15840
caggagcgcacactgcgcaatctttccctgtgggccaaggccctcgtggtgctggtcgcg15900
ctgacgagcatggacgacgtgctcattcggtacgtgcaacggcggggcgacgggctgttg15960
ctggacgagcagatcccgcgcgggaatgcgttcgccctggcagccgtcatcgtgctgttc16020
tgcgtcggcaggccgcgtcggcggatctgggcggcggctgctgtgctggccgtgccgacg16080
gtggctgccttgccgcagtggttcgaactctccccgcagcgcttcgtgtccgacgacgag16140
tgggcagtcacgttggcggcccagggggtcgccgcctgctgcatgctggctctcttgggg16200
ctcggcttcgtaactgccgcctggcgcttggccgttgacggggacctgctgccgatgagc16260
cgtcggcacccggggcacgcccgggtcctcaggctccgcatcgccgggccggtgatcctg16320
gtgtgtacggccgctgtggcgatctgtttcgccctggcccaggagcgcaactggcagcgt16380
gccacctggctcagcgatcgctcggaccccgcctacgcgaccggccagtggagcgatcgc16440
gtgtgggaggcggtgtggtccgtcgccaatgggcaggactggctctcgtggcaggcctgg16500
ctgctcacgggagttgcggtgcttgcggtcttgcgcacctggcgcgccccggcctccgtc16560
tcccctctggacgacccggcggaccgccttctgttcctcgccttcttcgccatcgtggcc16620
gcggcttccggcggctactttctgggcaacgaggtgctcaccggcttgtggattccgctc16680
agcatgctggctctctactgggtggtggttcccttcacccaccgctcggtactggcgcag16740
cctttcgagcggtccgggcggcccctcgccgattccgcggggcccggcgcacgcaccgta16800
ctgcttgccaaggcccgctcctaccgcgagacccatgccgaactgcgccgcctcgaccag16860
gggttgttcggggacgtgccaccgaagcgaagcgacctggaacaggagttgagcgacctg16920
cacaactggcccacggcaggtggctccgaccggcttcccgccaaggtgtccgtggtggac16980
ggagcactggcgctggggccacgagacacctggtgggccaatggcagccgctgtgcccgc17040
ctcgccttggttccggcggtaccggcggccctgctcctggcctgggtctggaaggtcaag17100
ggcgaggcctggcacgcgactctgcacgaacagttcggtctgccggatgtcctgctcttg17160
ttcgtcggggagatggtgatgttcaccagctcggcgttcgtcctgggcgcgctgtggcgc17220
catctgccagggcagcgcggcgccgccaaggccctgccggtgacactcgccttcgcgctg17280
cctatcggcttggacgcgctcgtctaccggttcaccggcgagagcaccgcgaacctcgct17340
ctggctgtgtcggcgatgctgttcgtgctgactgtcaccagcatcgctctcgacttcgac17400
acgttccgcggcgaacggcgttactggcagagccggttgggcctgctcctttcgatctat17460
cagatgcgttactactcgctgcaggccgcctacctgatcgcccaggtcgttgccatgatc17520
acgatctgggagttcttcgcggaacccgacgtggtgccgaagccctccgactcgaagtga17580
gccgggcgcaccctcccgtaggttacgggcgccactggtcctggctttgcggtagtcctt17640
ggtaggtgccgtagtccgttgagccttggtgggtgccgtagtccgttgagccttgctcca17700
tcgtggggtgctccgtcgtggagtgctccggcgtggggagctccgtcgtagcgtgctcca17760
tcgagccgatccaggcgcgcgccggggggcgtccgacgtacttgccgaagagcagtgccg17820
cggcggtggcagcgagtacgccggggaccatctcgtagacgcccgattccagcggcccga17880
gaagcgggtcgatgtacttccacaggaacacggtgagcgcacccgtcaccatgccggcca17940
tcgccccggctgccgtcatgcgcggccagaacagcgacaggatgatcaccgggccgaagg18000
ccgcaccgaatccggcccaggcgtacgcgacgatgtcgagcacggcgccgccgctcagcg18060
cgatcgcataggcgaccaatgccacggccaccacgctcagtcgtccgaccatcagcagca18120
acgtgtcggaggcccgccggttgaggaacgcccggtagaagtcctcggtgagggacgtgg18180
ccgagaccagcagctggctgtccaccgtggacttgatcgcggccagcacggccaccagca18240
ggattcccgcgatccaggggttgaccaggtgtgtggacagctcgatgtagacggtctccg18300
ggttgtccagcggctcgtcgagcacggcgatccccgcaagcccgatgagcgaggaacccc18360
ccagtacgacgaccacccagcccacacccagacggcgggccagcggtatgtcctttgtgc18420
tgcggatacccatgaagcggatcaggatgtggggttggccgaagtagccgagcccccagg18480
ccaacagcgagatcatcgcgatggcgccgagcggctcgccggccgaccacgtgttgccgg18540
cgaaggatgcctcggccaccgggtcgagtagtgccggggtcttgtcgctgagcgcgtcgt18600
gcagcgcgccgaagccgccgagccgccagagaccgagcgcggggaggacgagtgccgcga18660
ggaacatcagcgtgccctggatggagtgcgtgatgctcacggcccggaagccgccgagga18720
tggtgtaggcaacgatcaccacggcaaatacggtgagcccgaactcgaagtcggcgccga18780
atatctcgttgaacaggagaccgccggcgaccagcccgctggcgacgtagacggtgaaga18840
acaggaccgtgacgatggccgagagcagccggagcatcctgctccgatcctcgaaacgtt18900
cttccaggtacgacggcagggtcacggagttgccggccagctcggtgtaggtgcgcaagc18960
gaggtgcgacaaaccgccagttgagataggtgccgacgatcaggccgacggcgatccagg19020
tggcgccgatcccggccatgtacacggcgccgggcagacccagaaacaaccagccggaca19080
tgtcgctggcgccggcagacagggcggccatcggggcggtgagtcggcggccgccgaccg19140
tgaagtccgcgaatgtggccgtttccttttgcgtcatgacaccgatcatgaccatcgcaa19200
tcagaaagaccccgaaagtgatcatggctgggacggtcagggtgagcatgcattactccc19260
tgcaatgcgcggacgcgaccacctgcatgtcaagtacacatacaggccatcctcttcggg19320
ctgaagaggcggagtgtaggggcctcggcgcagaccggcgagagggtctcctgccgcctg19380
cgtacatcggttccggattgtgatccctgatgtttaccgcagatcgtgtacatgccaccc19440
caggtgggtaactacccgaccgcctccccccatgggccttgagcgatgcccggtgaagcg19500
gctgccccggcatgcccttcggtggtggcgctcctgttgacaggattcgtatgggcagtg19560
cacttcggctgggcagtgcagttcgacgatctcgtagacgtatggccgttgcctgtggcg19620
tggtcatcgccgtggctggtgggctcctgtgccccgttgtcgccgcgccgaccgccggtg19680
cggcggaccatgactttcggccgcagttggtgaccgtggacacccccacccgtgccgcca19740
aggagaaacttgccgggctcgggctcgacctgaccgagcatgccgggcatggctttgtcg19800
aagtcgtgctgcacagcccggccgacgcgctcgcgctgcaagtgggcggattcagctgga19860
aggttcgcgtacccgatctcgtccagcgtgagtccgacgtgaacgccgcgaaccgggcct19920
atgccgccgccaccggcacctcgccgctgccgtccgggcgggacagctaccgccggctcg19980
ccgactacaacgacgatctcggccggatggccgaccagaatcccggactcgtacggaagt20040
tcacgctcaagcacaagagcctcgaaggcaagcccgtgcacggggtggagatcacgcacg20100
acgtcacggctgtcgacgacgggcggcccgtcttcctgatgatgggcctgcaccacgccc20160
gcgaatggccctccggcgagcacgccatcgagttcgctcatgatctcgtcaggaactacg20220
ggagcgatgagcggatcacctcgctgctccagaaggcgcgggtgctcgtcgtgcccgtcg20280
tcaacgtcgacggctttgaaaagtccgtcaacgatgggcagttgatcgatctgcgggaga20340
tcgacgacggcggcaccggatcgatcctcgccacgcccggcaacgcctacaagcgcaaga20400
actgccggatcgtcgacggcctgagcccggtcgcgggcgagtgcgcgctggcgagcagcc20460
ccggcgggttcggtgccggtgtcgatctcaaccgcaactacggcggattctggggcggtc20520
ccggcgcggccgccgagtccgtgcaggccacgtaccgcggcgccgcgccgttctccgaac20580
cggagacgcagaacatccgcgagctggtcagcagccgccaggtgaccggcctgatcacca20640
accacaccttctccaacctggtgttgcggccgaacggggtcgcgcccgacacggtcggtc20700
cagacgggcagcccatcggcaacccgccggacgaggccgcactgaaggagctcggcgacc20760
ggatggccgagcagaacggctatacgagtcaacacagttgggagctgtacgacaccacgg20820
gcaccaccgaggactggtcgtacaacgcgacgggcggctacggatacaccttcgagatcg20880
ggccccacgagttccatccgccgttcccggaggtcgtcgacgagtacgtgggcgcgggcg20940
agtacgccgggaagggcaaccgtgaggctttcctgctcgccctcgagagtgccgtcgatc21000
ccgagtcgcactccgtgatcagtggcaaggctcctgccggggccacgctgcggctgaaga21060
agacgttcgccacgcccacctggtcgggcacgatcaaggacaccctcgacaccacgatga21120
ccgtcggcagcggcggcagctacacctggcacgtgaacccgtcgacccggccggtcgtca21180
aggcccgccagatcgaggtcatcggctccgagccgctgaagcggcagacctacacgggca21240
cgaccgcgcccggacagccgacggagcaggagttcgtcgtcgaccgggacgccgacgtct21300
tcgaagcgaagctcgactgggccacgcccgacgacctcgacctgtacgtcctgcgcaaga21360
acgccgacggcagcctcacccaggtcggcagttccgccggttccgtcggcgagaaggagc21420
gggtcctcctcgacgacccggagcagggtacgtacgtactccgcgtggagaactgggctt21480
ccgtcgcccccagttggaccctcaccgcgtccctctacgacgccaccgtggacgagatcg21540
gcggcgtcatcgagaactggacgctctcctgcgagaaggacggaaaggtgcttcagcagg21600
tgcccgtcgtcgtcgaccgtgggcagcgggtcaaggcggacttgaagaactgcgcgaagg21660
gctga21665
<210>2
<211>1872
<212>dna
<213>actinoplanessp.
<400>2
tcatagcgaagccgccacctccttgccgccgaccagctttcggcgatacgggtcaacccc60
gagggcgacgcttacgtagcgcccctcgtcctcgaatgccaggccgcgatcgacgaagta120
gcggagcatttcctcgagttccgcttccccgacgacgtgcccgctgtcggcaagccgccg180
gcgtatgccctcgcgggcggcgcactggaacatgccgaggtacacattgctgcggacctc240
gtccagctcgatcacttccgtcggccagctggcacggcggtcttcgatgacgacccggcc300
tcggtcatcggtccagtaggaaagggtgccctgcggataggccttggcccattcctcgca360
ggcctgcttcatctcgtcctcgatgggccctgagattccccggacgctggtgtcgaagaa420
gaacaccatgtcgtacagctgatcctgcgggatctggtagatgaagtcgtatatttccga480
ggggcggcggaacatgaacccctgggtggggtcctcgaagtagggactgaaccgctcaag540
ggctatgcgccaagccccggttggcggctccaggtgctcgagcgtggccaatttcttgag600
cagcccgcggtagtcgtcctcggtctcgcccgggaagccgtagaggatgctccatgtcac660
gttgagcccgagatcctgtccgtcacgcagcatccgtacgttgtgcgcggcactgacgcc720
cttgtccatgaggcgcagcacatggctgctcaggctctcgataccgggctgcacgaagag780
gacgttcgcctctttcagcctactcaactgctcccggttcatattggacttgatctcgta840
gtgaattcgcagatcgcagtcgagggcagctatctcgggcatggccgtattgagatactt900
catgtcgaggatgttgtccaccatgaccaggtcgaggatctggtgtcgctcggccagttc960
ccggacttcctgggcgatgcgctcaggggccttgctccggaagtcgatattcgatccgtt1020
caggccgcagaacgtgcattggtgagcctctccccaccagcaaccacgggaggtctcaag1080
gaccagcatcggacggacgtggtgacggacgggtgacctttcgagggcctgaaagtagct1140
gtcgtaaccgggcgcgggcaccatggcgaacggcagcgccgccgtggccggtggattcac1200
caccggatgcccgtcatcccccctccagctgagccccggcacgtcggcgaggctctcgcc1260
ccggatgatgcgattcagcaacgcgggcagcgcacgttcgccctcaccgctgatcacgaa1320
gtcgagttgctcgaaattccggtgcaacgcgggaccttgtgctccgtcgcagttgctgcc1380
gccaaggaccgtgcggatgcccggcgcgagtttcttcagctccctggccagtgcgagcga1440
cgggacgttctgcatgaaggtgctcgtgaacccgaccacgtcgggaggatcggcagcgat1500
ctcggccgcgagatcccggatgaatccccgggcgtacttgtgcatctcaacgggaagtgt1560
cgggtccatgtcccgctgctcgaggaacttcgcgtactcgtcgacctgataactgtcgac1620
gtcgtacagcgctggggtgaacacccagtcccctacgccgtggaagacttgatccgcgat1680
gttcccgtagtcctcgcaggtgacggagccgttgctctcccgcatcaggtattcggccca1740
gcggaggttggcgtacagctcatcgacggtccagtcggcggcgttcttgcggacgcatgg1800
ccccagtacgcccagcgcgctggacggcgtgtcgagcccttgccacggcatggcgatcat1860
caggagtttcac1872
<210>3
<211>1089
<212>dna
<213>actinoplanessp.
<400>3
tcagaaccagttcccacggtccgcccggccgagcgccgccaacagcgtccgatgtgcctc60
ggcctctcccactgagatccgtacaccgtgccccggaaaggctcggaccttgacccctgc120
ggtagccgcagtccgggcgaaagactcggcggccgaagcgagcgggagccagacgaagtt180
ggctcgggaaagcaggacgggcagcctcagttccctgagttccgcggtcagttcttcgcg240
tgccgcagccactgctgccagacgttcacacagttcgtcctcgctgcgcagcgagagcat300
tgcggcttgttccgcgaagcgcgtcactccgaaagggattgccgtcttgcggacggtggc360
catgacctgccgtggcccggccgcgtaaccgacccgtaggccggcaaggccataggcctt420
ggagaacgttcgaagtaccacggtgttgctgtgctcgctcagcaacaccggcagacccgg480
aggattggcgccccggtcgaactccacgtacgcctcgtcgaggaccgcgaccacatgagc540
cggcagcgaacgcaggaaaccgtgcagctcgtcttggtcaatcacggttccggtcggatt600
gtgcggggagcacaggatcaccaccctggtccgcgcattcacccgggtgcggatctcatc660
gagatcgtggccgccggacgcagtcaggggcacgtggactccggtggcacctgaaatggc720
gaccaacagcggataggcatcgaatcccggccagccatggacgacttcgtcgcccttgcc780
gcacagtgcgagaaggatctgctggagcacgcccgcgcttccggggccgaccgcgacctc840
atccggggagacgcacaagtgcccggcaatgtcctcggtcaggtcccgtgctgtggggtc900
ggggtaacgagcaagtcgcggcaagcctttttcgataccggcaagcacggtaggcagcgg960
ggggagaaccagctcgttgctggacaggtcgaaggtgaaccgcgagctgccttcggcgtt1020
cgacgactccttgtcccggtaggcccgcatgtcccgcagggtgctgcgttctgcgaatct1080
cacgttcac1089
<210>4
<211>957
<212>dna
<213>actinoplanessp.
<400>4
tcacgcctgcagactcctcgccgtgcgtgattcgcgggtggcgagcagctcgtagagcat60
ttcgtggaccggcgcggtcaggcccgccccgcgggcgaggcggaccacggccccggtcca120
cgcttcgagctctgacggccgtcccgccaggatgtcccgttgcagcgaggaggtgacgtc180
gggcgactgctggtccatgagctctgtcgcggtgtccacggcagctgccggcagcgcgat240
tccgagcttgatcccggtctcgtagatctcccgcatgccggcgatcagaatgttgcgggt300
gccggtgcgcgaccggagctcgccgatggtcgccccgccggtggcggctccaaggctgcc360
gatcgggaccaccaacaggaacttcgcccaaaggccggcccagatgtcgctcggctcggg420
cacggacaccgaggcagcacgcagcacctcgcgcagtcgtgccacccggtcggacacagt480
gctgtcccactcggtgaaggccagagcgccggggggacccacgtgcctcaactcgcccgg540
accggccgtcgaggccacgaccctgacgctgccggggagtacccgaccgcggccgatcct600
ggctgcgacctgctcaggggcttccaccccgttctgcaccgtgaccacggcagtgtgctc660
gccgaccagcgggcccagcgcgtcgagggccgccggcagctgtgaggtcttgacgcagag720
cagtacgaagtcgacctcgccgatgtccttcgggtcggccgacgcccgaacgtccggcac780
acgtaagtcacttgagccgttggtgatgcgcagcccctgtcgcctgagcgcggcgaggtt840
ctcgccgcgggccaggaaccgcacatcatgcccggcggcggcgagcaggccgccgaaata900
gccgcctactccgccggctcccaccacagcaatgctcggaccgccttgttccgtcat957
<210>5
<211>1215
<212>dna
<213>actinoplanessp.
<400>5
tcatcgcggcaacctctctcccactcgtatcgggagggcctcgaggccccggctgagcga60
actgtcggagagccaggcgacctcttccggaggcacggcgagctcgaatcgcggcagtcg120
gcgcaacagcgtggagaaggcgatctcgccctggagtcggcccagcggagcgccgatgca180
gaagtggggaccgtggccgaaggacagatggcggttcggtgaccgcgtcacgtcgaaacc240
gtccgggtcgtcgtacacgtcgggatcacgacccgcgctggacagcgacaggtgtacgaa300
gctccccttgggtatgtccactcccgcgagctgcatgtcctccgccgcgacccgcagcga360
ggcccaggcggccgacccttcgtagcgcaggatctcctcgatcgccgaagggatcagctc420
gggggtggctcggagcatctcgagctgctggggatggcggagcaacagtgctgtgccgtt480
gccgatcatgttggcgaccgtcttgtgaccggcgatgatcagcaggaggagcgtcgaaag540
gagctctgtttcgctgtatacgcctgcgtccctggccctgatgatctcgctcagcaggtc600
gtcccgcaggtccgtgcgccgctcggccacgagcttggtgaaatagtccgtgaactcttc660
gctcgcggctttcagctccgcctcgtcgtgctgcaacgggtcctggctgaggatgtagct720
ccactccagaaacagcggccggtccgccaccggtatccccaggtattcgcagatgacggt780
gagcggcatcggcagagcgaaggaactgagcaggtcgatttcgccgttctccggaaaggt840
gtcgatgagatcgtcgacgatgtcctggatgcgcgggcgtagttgggcgaccgtggccgg900
caggaatgcccggctgatgggcttcttcaggcgggtgtgtttgggcgcgtcggcgaatcc960
caggttgccgaccacgaggagactgctggccacggccttgtctcgatagcgggcgggcag1020
gttctcgacttgcttggagaggcgcgagtcgccgagcgcttcctcaacgactttgttccc1080
gaggaccgcgtacgcgtcggcgccggggggaacattgatccgatggaccggacacttgga1140
cctgtactccgcggctgtcgcgtgcggattcgaaccgggctcggtgaagaattccgtggg1200
aatcacgtcggtcat1215
<210>6
<211>303
<212>dna
<213>actinoplanessp.
<400>6
tcatcgtgactcctcggctcgcgcggcttcgctgccacctgctaccgccggcactatcca60
gatgacgtcgtggtgctccactttcgtttcgaggccgtcgaggctcctgatgtcgctgtc120
gttccggtagacattgacgtagcgcttcacggacccttcctggtccatgagtcgctcgag180
gacccccggacaggtctggtcgagaccgacaaggacctcccggatattggcgccctcgac240
aggcaactgccgccgaccgccggtcaggacgtggaaggctgcgggaagtttgacatcggg300
cat303
<210>7
<211>717
<212>dna
<213>actinoplanessp.
<400>7
ttattcctcctcgagtaggacttcgaactgctcatggatcgccgccggaatccgtatgtc60
aagcctgtcgcattgctcggcaccgggccgccaggccgctacccgatgtgcgataccgcc120
actactctcatgcacgtaccagatgacgttcactggccagcatgactgccggaaaaggta180
ttcatccatctgtgtcggatgttcgctcaggcgctggcgccgttcgtgcggcgggccgat240
ttcatgccagttcggatgtgaatgtatggagcccagcaactccaggccgttcgccgactg300
ttgcctgatcgcctgcaagacgccctgttcatcggaccagaatcctcgccccggattctt360
gtacacgtcgccgaactgcggggcgatcgtcgcttcgaattcggccatgacactctcgtc420
actgtcccggacattcggcacgaattccacatcgctgatcactatttcagcgccgcccac480
ctgcccgaacaacagacccgatgcacgcggcagatggctcgggacgccatcggcgtcgcg540
gctgtccaggcatttttggtactcccccagtgcgctgctcaggaaacgcctgacgggttc600
gatttcaaatcggacggtcaccggcagatcgtcgagcagggctgcactgtctgctgtgcc660
acccagaccgtcatccgtagcagctgtcacatactgggcatctttcacgcctgccat717
<210>8
<211>1491
<212>dna
<213>actinoplanessp.
<400>8
tcatgggcgtacgccccctgcgctctggcgaggcaggtcagtcacccctggcatacgagg60
gccgcggagcgtggaggtcttgatgccgacccagcacaacagccctccggccgccgctac120
ggcggcagcgatccagaaggcggtggtgaagccgtcgcgcgccggcgtatcggaaccgga180
gatatgccaggcagccagcaaggcgacagccagctgactgcctacgacgccgcctgccgt240
gcgcaccaccgtgttgacgccattggcggttgcggtgcgtcgggcctccgtcaagtcgct300
gatcaccgacgggagtccgctcagcaccagtccggaaccgagacctacgaccaggtaacc360
gaccgccacttgccacccgttggcattccacgcccagagcgagatcgccccgaccgccat420
cacggcgaagcctgatgcgagcgtcgcgcgtacggacgtcaggcgctgcagcagacctgc480
tagtggtcccgcaggcaacagcaccaatgagccgggcagaaggagcagacccgccatggt540
gacatcggccccgagcccgtagccgatggtccctccgcccggtagccgctgatccgccgc600
ggtctgggcgtacgtcggcagcaggacgtagaacacgaacgagaccacgccgaacacgaa660
cgccgcaccgtgcaccgagacgaaggagcgaccggccacgaccgcggggtcgatcaaggg720
ggcgggcgactttcgttcgactacgaccagcaatcccaggagaaccgcggaggccccgaa780
cagagccagcgttcctgtggacgcccacccccatgaggtgcccttggtcaacgcgagcaa840
cagcgcgacgaggaccaaggccaggagcaccgccccgggcacgtccaccggttcgccggc900
ttcgccccgctggtccggaacgtacttcgcaacgagcccgatcgcgcccaggatcagtac960
cgcggcgacggcgaacagccagcgccatgactggtggtcgacgaccagacctcccacgac1020
caggccgatgccggcccccacaccgatggtccctgacaccagcccgaggcccgaacgcag1080
ccgctgctcaggaagtacgtcgcgcaggatgccgaaggacagggggatggccgcgaggct1140
gactccctgcacggcacggcatgcgatcagaaccccgatattgcctgccacagcacaacc1200
gaccgtgccgatcaggtaggtggtgagaaccagcagcaaaactttgcgcttgctgtagcg1260
gtcccccagccggctcagcaagggcgtgctcgccgcactggtcagcaggaacacactcag1320
gatccacgccgaccacgtcgaggccgtgtgtaactgcacctgtagtacgtgcaaggccgg1380
gacgaccatggtctgcatgagtgcgtacgacagcacggagacgccggtggcgatcagcgt1440
catcgtcgcgctgatcgacggtggacgtggcgaagtgccgttgtaggacaa1491
<210>9
<211>942
<212>dna
<213>actinoplanessp.
<400>9
atggaactggatttgcggcacctcaggtacttcgttgccgtagccgaggaaggcgggttc60
acgcgagccgcggcccgcctgcacatgacacagccgccgttgagcgtggcgattcgtcaa120
ctcgaaagagagctgggtctccagcttttggacagaacgggcaacagagtcgaactcacg180
tcggtcgggcgcgacttcctgactcacgcgaggaacttgttgcagcagtggcaggtcacg240
gtcgagaggatgcggcaggcggggtcgcaggatgtcgaacggctcgtcgtcgcgttccgc300
ccggccgtcagccgccctctggcacaccggaccattgaactcatccgcgaaaagcaccct360
gagtatcaggtagtgccccggtacgtaccgtggaccgaacagacagcatgcctggaggca420
ggggacgctgacgtgtccttcgtgctggagcccgcggactacgtgggcctcgagagggcc480
accgtggccctgttaccccgggtcgtctgtctgccatcggctcacgagctggccagtcgt540
gactccgtgtcgatcgacgacctgagcgaggttccgatcattcgccccaccggcgggtcg600
cccgagtggtccgacttctggggtggtgaggtgtgccccggcaagcgcacctggaaggaa660
cctcccacagcgacgcgcctcgacgaggccatcgacctcgtggccctcgagaacgcagcc720
gcgctcgtccccgtctctgtcatggcagtccagcaccgtcaggacgtcgtcttcatccct780
gtgacggatgtgcctgccgcccggttgtcccttgcctggcgtgagggttccgactccgaa840
ctggtacgcctcgccgtcaggtgcgctcaggccgcagcccaggatccagccgtcaggacg900
ctcttcggagaacctcgaccaaccggaaccgctccggcctga942
<210>10
<211>903
<212>dna
<213>actinoplanessp.
<400>10
gtgcgtcgttgggcagcacagtcgggccccgaagacctgtatttcgtcagcaacctgcac60
gccatgacgaccaagcacgaccccgaacgtctccaggaactgaccgaccaccaactcgct120
ttactcatcgcggcgggcgtaccccaggaacgtctcttcgtgcagtcggacctcatccag180
gagcacatggcgttgacgtggcttctcgagtgcacctgcaccttcggggaggctcgaagg240
atggtgcagttcaaggagaagtcccaggggagcaactccgtacgccttggtctgctcacc300
taccccgtcctcatggcggcggacatcctgcttcatggcgcttcagaggtgcccgtcggt360
cacgatcagaaccagcatgtggagctggcccggaccttggcgcggcggttcaacacggac420
tacggcgaggtgttcacggttccgcaagccgtcctgcccgtagccgcagcccgggtacgt480
gatctcgctgcccctacgcggaagatgtcgaagtcgtcctcggacggcagcggcatcgtc540
tacgtcctggacagcccggaggccgtacgccggaagttccaacgcgcagtgacagacgga600
gaaaacaccgtccgctacgccccggacgaacagccgggcgttgccaacctcctggagatc660
agggctgcctgcactgacacgctcccgagcgatgcggcgaagggtatcgattcctaccgt720
gacctcaaggaagcagccgcagaggcagtgatctccctgatcgcaccggtgcgtgagcgg780
gcactgcagctcctcgaagagcgatcggagctggcgaagatccgggctgagggggccgac840
cgtgctcgggcgcggtcacgagaccgcttggatcgtgcgctcagccttgccggtctgaag900
tag903
<210>11
<211>795
<212>dna
<213>actinoplanessp.
<400>11
atggctgacgatcccctggtcatcggtggtacgagctactcgtcgcggctcatcatgggc60
accggcggcgcccccagcctggacgtgttggaacggtccctggtggcgtccggcaccgaa120
ctgaccaccgtcgcgatgcgccgcgtcgacccgagcgtgaagggctcggtgctctccgtc180
ctcgaccggctcggcatccaggtgctgcccaacaccgcgggctgtttcaccgcgggcgag240
gccgtcctgacggcccgcctggcccgcgaggcgctcggcaccgacctggtcaagctggag300
gtcatcgccgacgagcggaccctcctgcccgatccgatcgagaccctggaggcggccgag360
acgctggtcgacgacggcttcacggtgctgccgtacaccaatgacgacccggtgctcgcc420
cgcaagctgcaggacgtgggctgcgcggcgatcatgccgctcggctcccccatcggctcg480
ggcctcggcatccgcaacccgcacaacttccagctgatcgtggagcacgcgtgcgtgccg540
gtgattctggacgcgggtgcgggtacggcgtccgacgcggcgctcgccatggagctgggc600
tgcgccgcggtgatgctggcctcggcggtcacgcgcgcgcaggagccggtcctgatggcc660
gaggggatgcggcacgcggtggaggcggggcggctcgctcatcgcgcgggccggattccg720
cgccgccacttcgcggaggcgtcctcgccgaccgagggcatggcccggctcgacccggaa780
cgtccagccttctga795
<210>12
<211>1179
<212>dna
<213>actinoplanessp.
<400>12
gtgtcgctgccacccctggtcgagccagctgctgagctcaccgtcgacgaggtccgcagg60
tactcccgccacctgatcatcccggacgtcgggatggacgggcagaagcggctgaagaac120
gccaaggtgctctgtgtgggcgcgggcggcctgggctcgcccgcgctgatgtacctggcc180
gccgccggcgtcggcacgctcggcatcgtggagttcgacgaggtcgacgagtcgaacctg240
cagcgccagatcatccacagccaggccgacatcggccgctccaaggccgagtcggcgaag300
gactcggtcctcggcatcaacccgtacgtgaacgtgatcctgcacgaagagcggctcgag360
gccgagaacgtgatggacatcttcagccagtacgacctgatcgtcgacggcacggacaac420
ttcgccacgcgttacctcgtcaacgacgcctgcgtgctgctcaacaagccgtacgtctgg480
ggctcgatctaccgcttcgacggccaggcgtccgtcttctggagcgagcacggcccctgc540
taccgctgcctctacccggagcccccgccgcccggcatggttccgtcctgcgccgagggc600
ggcgtgctcggcgtgctctgcgcgtcgatcggctccatccaggtcaacgaggccatcaag660
ctcctcgcgggcatcggcgacccgctggtcggccgcctgatgatctacgacgccctggag720
atgcagtaccgccaggtcaaggtccgcaaggacccgaactgcgcggtgtgcggcgagaac780
cccacggtcaccgagctcatcgactacgaggcgttctgcggcgtcgtctccgaggaggcc840
caggaggccgcgctcggctccacgatcactccgaagcagctcaaggagtggatcgacgac900
ggcgagaacatcgacatcatcgacgtccgcgagcagaacgagtacgagatcgtctcgatc960
cccggcgcccggctgatcccgaagaacgagttcctgatgggcggcgccctgcaggacctg1020
ccgcaggacaagaagatcgtcttgcattgcaagacgggtgtccgcagtgcggaagtcctc1080
gcggtcctgaagtctgcgggcttcgccgatgctgtgcacgtgggtggcggcgtgatcggt1140
tgggtcaaccagatcgagccgagcaagccggtgtactag1179
<210>13
<211>1275
<212>dna
<213>actinoplanessp.
<400>13
tcagcttgtcgtggcggtggcgccggcggttccgccgctgaccagcttggcccaggtggc60
ggggccggcgatgccgtcggaggtcaggcccttggccttctggaaggcggtgagcttgct120
ggtggtggccgggccgaagacgccgtcggcggtgacgtcgtagccgttgtcggcgagctg180
gcgctggagggcggtgacgtcggtgcccttcgagcccgacttcaccgtggcgatcagctt240
cgcccaggtggcgggaccgaccatgccgtcggcggcaaggccctcggccttctggaacgc300
ctggaccttcgcggctgtccccgtaccgaacacgccgtcagcagtcgtcgcgtacccgtg360
cgcaccgagcagcaactgcacggtcgccacgtcgacgcccttgtcgcccgccttcaccgt420
cggccacgacgtgcccggctgcggctgcggcgcaccgcctcccttggcgagggcccgcag480
ctggtccaggctgccccggtacacgttgcggtcgcccttgcccgcggcgccgttcttgcc540
ggggaggcctgggatcgcctcggactccgtgtactgccagagcgaccaggcacccgcgcc600
cggcacgtcctgcggctccttcgacccgctttcgtagcgggccagccacagcgggtggtc660
cttgaagacctggcccttgccggccatgcagccgttcacgaacgacgcccgtgtgtagac720
gatcggcgtcaccttgaacgcctcctccacgcggttcaggaaggcggtgagctggtcggc780
gcggagcgccttcgggcacacctccttgccgttcacccacgtgccctcgacgtccagaac840
cggcggcagctccccggccctcttgccggtgtagccggcggaccgggccgcgcggatgaa900
gtggtcggcctgcgcgccgccgtccgtggtgctcttcgggtcgaagaagtggtacggggc960
gcgcagcagcgatgtgccggacgcgtccttgaagtcccgtgcgaaccaagggtccttgta1020
accggtgccctgcgtcgccttgaggaacgcgaaggagttggactgggcgacgcgcttcca1080
gtcgatgggcttgccggtcgcgtcgtggttgtggtggctggtgtcgacgcccttgacctc1140
gtacgtactgggaggggcggccacgctctcgtccgcggtggacagcaggacaccactcat1200
gagcgacgcggtggcgacggccgccgcggccaggcgcagtccgtgacgacggccgcgcgg1260
gttctccatggacac1275
<210>14
<211>996
<212>dna
<213>actinoplanessp.
<400>14
atgggcgttgaacgcgacgagccgacgcggtcagcgaggcgggaactcgccctgctgttg60
cggagctggtgggaggcgcaccccgacaagatcacgcaggaggcgctggcccggcggatc120
acggagcggggcgtacggatcagccaggagatgctgtcgcgctacctgaaccggtcccgc180
ccgaccacggcccggcccgacgtgatccgcaccatgcacgaggtgctgcgccgggcgccg240
gaagagctggacgtggccctggaactgcacgctcgggccaccgccccgcagacgccgccc300
gccgagggggcggccacgagccagccggccggggacgcggggaccgccgcgccgaagggc360
gtagagccgacctcggcggccccgctcttgacccgcacgccccacacgccccgccccgct420
tcgcggaagaagtggccgtggatcgccgtcgtcgcggccgcggtcgtcggcgcgtccggg480
ctcaccgccttcatgacactgggcgaccagcggcagaacaccccgcggggacacggagcg540
acaccctccgcctcacccaccgccctggtgtcacccaccgcccaggggtcgcccgccggc600
acgcatcctcccgcggagtgccgcgacgagtcctgcttcggcatcgacgccaagtacgcc660
atctgccaggacgacgccgccacttactacacgggccgcgcccacggcgtcctcgtcgag720
ctgcggttcagccccgcctgccaggcggcttgggccaagatgagcggcacctcgcagggc780
gatgtcgtacgcgtcaccaacaacgcgggccgcagccgccactacacccagcagtggggc840
cgcgacgcccacaccacgatggtggaggccgtgagccccgacgacgccaaggcttgcgcc900
cgcaccccgcgcggcgaggtgtgcgccacgaaggccgtcgcgtccgccccgcgcgacgcg960
gcacctggcgagcgcgcggcacctggcgggcgctga996
<210>15
<211>1041
<212>dna
<213>actinoplanessp.
<400>15
atgcccgcacgcaccacacgaaccgcacacaccacacgcaccggccggttggccgtcgtc60
gccctcgcggccttgacctgtgcgggcctggtcaccggaactgcagccacggccaccaca120
cccgactccctgcccaccgcgaagcgcgccgcagcgcccgacgcagcggctgtatcgtgg180
ccgacgctgaaggcgggcgcgcgcggtacggaggtgaccgcgctccagcacctgctgatc240
gcccgcggccaatccgtcgccgtggacggggagttcggcccggccaccaccacggccgtc300
aaggcgttccagaaggccgacgggctcaccgccgacggcatcgtcggacccgccacctgg360
gccaagctcgtcccgacgctgcgtcagggcgcgcagggcgcggcggtgaaggcggcccag420
accctgctgaagacccgtggccaatccgtcgccgtggacggggagttcggttcggccacc480
acctcagccgtcaaggcgttccagaaggccaaggggctcagcgccgacggtgttgtcggc540
acgcagagctggtccgcgctcctcacctcggactccggcgcgccgtccgggaaccgggcc600
gcgttcgcccagcagatcctcaacaccagcggcatcgagctggcgaccgtccaccccggc660
ggcacccacgccggctccaccgcccggcagaacatcatcgacacagccaacggcaagggc720
gctctgaccagtccctggagcgacaagccgaaccagcgcgtggcgctcgacacccggatg780
ctcaacgggctgctgaagctgctctcccaggacggctaccggatctctgtctccgagatc840
gtcggcggcgaccacagcacgaactcccggcactacgcgggactcggcttcgacatcaac900
tacatcaacggccggcacgtcggcgagagcgccccgcaccagggcttgatggccgcgtgc960
cggaagctcggggccaccgaggtgctcggtccgggcgacgccggccacagccgccacgtc1020
cactgcggctggccgcgctga1041
<210>16
<211>723
<212>dna
<213>actinoplanessp.
<400>16
ctactccgtagcaccgtccgatttcttgccggctgcggccaaggttcctcccaatagggc60
cgcgcccaggggccatgaacgcatgtgcacggggagggtcacgcagtgatccccgacgcg120
catcacaaggcccttgttcaccaaatcgcgtgcggcggcggcgaattgcgagtttcgggt180
cacctcggcccacgcgcatccctccaggcccaggaggaagacggccatctgggtgggatc240
gtccacgatgatttcgcggctggattcggggcgctgatcgacgacggacaggaacttcgg300
gcccttacggaagtagaggaggccgaaattgctggaggaccgccactcaccgacggacgg360
atgccctgtctccagcaccgtgatgctgtcgggggcaggaaggtgatcgagacgggggac420
caggtcgagctgttcggcgcccagggtcagtgaccaggtgactctggtgccgatcgagga480
acattcgcgaatcaacgcgatgatccgcacgatcacgtgactggggagtgccgagaagtc540
ggcgggctccggcagccgtaccccggcgacgggcacccggtgcagtagcgccgccagttc600
ggtggcgacgggaagttcggcccggtcgagtccgagggtcagcgtctcggtcgcgggacg660
gccaccggccggtacccgtccctcgtccgaatctcgggccttgagttcatcgatgtccag720
caa723
<210>17
<211>2541
<212>dna
<213>actinoplanessp.
<400>17
gtgtcggcggtgctgggcgttcttctcgcgatatcactcgcaactgcccccgcccatgcg60
gcagttcgctcggccgcggcggtcgatgtctgtcggtcggccgccctgagcaaggcgcgt120
gtgagcacgtgggtgcggcttgagcaccgcgatggtacgtacagcaggatccgcagcgag180
ctcagcgtcgaggtgcccgaggattggccgttggccaaggacctgctgctgagtgaggac240
agccgccggtacgtcgcggcgatgtcctgcctcacccgtaccgatcggggccggcaacgc300
cgctggtcggagtggaggagcagccgtccgacggtggcgtccacgaagagcggtggggtg360
aaggtcgtcgaccgtacgcactcctgggtcaacgtgtatcgggcgcacatcgatgtgggt420
acctggcgggtccgtgcgggtgcggagcgctggaccgtacaactgcaagctccgtccgcg480
ctgaacgcggcccgctgggatgagatcagggtggaacccggcgccccgggagccgagtcg540
gcgaccccgcggcctgacgaggggcgcggcgccacggcgttggtgtggcatccccagaac600
caccgtgagaaggcggctgctcctgccgtgagcgttgcgctcaagccctcctggcagcgt660
tcgtgggcagcccagaacgaccggctggtcgccgtggcgctggatcggggcggatggctg720
ctctgggacgcgacgagtgccgccctgttgctgtacgcaaccgtcctgtaccggaggcgt780
tccgctcctcccactcaggctcaggagcgcacactgcgcaatctttccctgtgggccaag840
gccctcgtggtgctggtcgcgctgacgagcatggacgacgtgctcattcggtacgtgcaa900
cggcggggcgacgggctgttgctggacgagcagatcccgcgcgggaatgcgttcgccctg960
gcagccgtcatcgtgctgttctgcgtcggcaggccgcgtcggcggatctgggcggcggct1020
gctgtgctggccgtgccgacggtggctgccttgccgcagtggttcgaactctccccgcag1080
cgcttcgtgtccgacgacgagtgggcagtcacgttggcggcccagggggtcgccgcctgc1140
tgcatgctggctctcttggggctcggcttcgtaactgccgcctggcgcttggccgttgac1200
ggggacctgctgccgatgagccgtcggcacccggggcacgcccgggtcctcaggctccgc1260
atcgccgggccggtgatcctggtgtgtacggccgctgtggcgatctgtttcgccctggcc1320
caggagcgcaactggcagcgtgccacctggctcagcgatcgctcggaccccgcctacgcg1380
accggccagtggagcgatcgcgtgtgggaggcggtgtggtccgtcgccaatgggcaggac1440
tggctctcgtggcaggcctggctgctcacgggagttgcggtgcttgcggtcttgcgcacc1500
tggcgcgccccggcctccgtctcccctctggacgacccggcggaccgccttctgttcctc1560
gccttcttcgccatcgtggccgcggcttccggcggctactttctgggcaacgaggtgctc1620
accggcttgtggattccgctcagcatgctggctctctactgggtggtggttcccttcacc1680
caccgctcggtactggcgcagcctttcgagcggtccgggcggcccctcgccgattccgcg1740
gggcccggcgcacgcaccgtactgcttgccaaggcccgctcctaccgcgagacccatgcc1800
gaactgcgccgcctcgaccaggggttgttcggggacgtgccaccgaagcgaagcgacctg1860
gaacaggagttgagcgacctgcacaactggcccacggcaggtggctccgaccggcttccc1920
gccaaggtgtccgtggtggacggagcactggcgctggggccacgagacacctggtgggcc1980
aatggcagccgctgtgcccgcctcgccttggttccggcggtaccggcggccctgctcctg2040
gcctgggtctggaaggtcaagggcgaggcctggcacgcgactctgcacgaacagttcggt2100
ctgccggatgtcctgctcttgttcgtcggggagatggtgatgttcaccagctcggcgttc2160
gtcctgggcgcgctgtggcgccatctgccagggcagcgcggcgccgccaaggccctgccg2220
gtgacactcgccttcgcgctgcctatcggcttggacgcgctcgtctaccggttcaccggc2280
gagagcaccgcgaacctcgctctggctgtgtcggcgatgctgttcgtgctgactgtcacc2340
agcatcgctctcgacttcgacacgttccgcggcgaacggcgttactggcagagccggttg2400
ggcctgctcctttcgatctatcagatgcgttactactcgctgcaggccgcctacctgatc2460
gcccaggtcgttgccatgatcacgatctgggagttcttcgcggaacccgacgtggtgccg2520
aagccctccgactcgaagtga2541
<210>18
<211>1647
<212>dna
<213>actinoplanessp.
<400>18
ttacgggcgccactggtcctggctttgcggtagtccttggtaggtgccgtagtccgttga60
gccttggtgggtgccgtagtccgttgagccttgctccatcgtggggtgctccgtcgtgga120
gtgctccggcgtggggagctccgtcgtagcgtgctccatcgagccgatccaggcgcgcgc180
cggggggcgtccgacgtacttgccgaagagcagtgccgcggcggtggcagcgagtacgcc240
ggggaccatctcgtagacgcccgattccagcggcccgagaagcgggtcgatgtacttcca300
caggaacacggtgagcgcacccgtcaccatgccggccatcgccccggctgccgtcatgcg360
cggccagaacagcgacaggatgatcaccgggccgaaggccgcaccgaatccggcccaggc420
gtacgcgacgatgtcgagcacggcgccgccgctcagcgcgatcgcataggcgaccaatgc480
cacggccaccacgctcagtcgtccgaccatcagcagcaacgtgtcggaggcccgccggtt540
gaggaacgcccggtagaagtcctcggtgagggacgtggccgagaccagcagctggctgtc600
caccgtggacttgatcgcggccagcacggccaccagcaggattcccgcgatccaggggtt660
gaccaggtgtgtggacagctcgatgtagacggtctccgggttgtccagcggctcgtcgag720
cacggcgatccccgcaagcccgatgagcgaggaaccccccagtacgacgaccacccagcc780
cacacccagacggcgggccagcggtatgtcctttgtgctgcggatacccatgaagcggat840
caggatgtggggttggccgaagtagccgagcccccaggccaacagcgagatcatcgcgat900
ggcgccgagcggctcgccggccgaccacgtgttgccggcgaaggatgcctcggccaccgg960
gtcgagtagtgccggggtcttgtcgctgagcgcgtcgtgcagcgcgccgaagccgccgag1020
ccgccagagaccgagcgcggggaggacgagtgccgcgaggaacatcagcgtgccctggat1080
ggagtgcgtgatgctcacggcccggaagccgccgaggatggtgtaggcaacgatcaccac1140
ggcaaatacggtgagcccgaactcgaagtcggcgccgaatatctcgttgaacaggagacc1200
gccggcgaccagcccgctggcgacgtagacggtgaagaacaggaccgtgacgatggccga1260
gagcagccggagcatcctgctccgatcctcgaaacgttcttccaggtacgacggcagggt1320
cacggagttgccggccagctcggtgtaggtgcgcaagcgaggtgcgacaaaccgccagtt1380
gagataggtgccgacgatcaggccgacggcgatccaggtggcgccgatcccggccatgta1440
cacggcgccgggcagacccagaaacaaccagccggacatgtcgctggcgccggcagacag1500
ggcggccatcggggcggtgagtcggcggccgccgaccgtgaagtccgcgaatgtggccgt1560
ttccttttgcgtcatgacaccgatcatgaccatcgcaatcagaaagaccccgaaagtgat1620
catggctgggacggtcagggtgagcat1647
<210>19
<211>240
<212>dna
<213>actinoplanessp.
<400>19
ctacccgaccgcctccccccatgggccttgagcgatgcccggtgaagcggctgccccggc60
atgcccttcggtggtggcgctcctgttgacaggattcgtatgggcagtgcacttcggctg120
ggcagtgcagttcgacgatctcgtagacgtatggccgttgcctgtggcgtggtcatcgcc180
gtggctggtgggctcctgtgccccgttgtcgccgcgccgaccgccggtgcggcggaccat240
<210>20
<211>1956
<212>dna
<213>actinoplanessp.
<400>20
gtgaccgtggacacccccacccgtgccgccaaggagaaacttgccgggctcgggctcgac60
ctgaccgagcatgccgggcatggctttgtcgaagtcgtgctgcacagcccggccgacgcg120
ctcgcgctgcaagtgggcggattcagctggaaggttcgcgtacccgatctcgtccagcgt180
gagtccgacgtgaacgccgcgaaccgggcctatgccgccgccaccggcacctcgccgctg240
ccgtccgggcgggacagctaccgccggctcgccgactacaacgacgatctcggccggatg300
gccgaccagaatcccggactcgtacggaagttcacgctcaagcacaagagcctcgaaggc360
aagcccgtgcacggggtggagatcacgcacgacgtcacggctgtcgacgacgggcggccc420
gtcttcctgatgatgggcctgcaccacgcccgcgaatggccctccggcgagcacgccatc480
gagttcgctcatgatctcgtcaggaactacgggagcgatgagcggatcacctcgctgctc540
cagaaggcgcgggtgctcgtcgtgcccgtcgtcaacgtcgacggctttgaaaagtccgtc600
aacgatgggcagttgatcgatctgcgggagatcgacgacggcggcaccggatcgatcctc660
gccacgcccggcaacgcctacaagcgcaagaactgccggatcgtcgacggcctgagcccg720
gtcgcgggcgagtgcgcgctggcgagcagccccggcgggttcggtgccggtgtcgatctc780
aaccgcaactacggcggattctggggcggtcccggcgcggccgccgagtccgtgcaggcc840
acgtaccgcggcgccgcgccgttctccgaaccggagacgcagaacatccgcgagctggtc900
agcagccgccaggtgaccggcctgatcaccaaccacaccttctccaacctggtgttgcgg960
ccgaacggggtcgcgcccgacacggtcggtccagacgggcagcccatcggcaacccgccg1020
gacgaggccgcactgaaggagctcggcgaccggatggccgagcagaacggctatacgagt1080
caacacagttgggagctgtacgacaccacgggcaccaccgaggactggtcgtacaacgcg1140
acgggcggctacggatacaccttcgagatcgggccccacgagttccatccgccgttcccg1200
gaggtcgtcgacgagtacgtgggcgcgggcgagtacgccgggaagggcaaccgtgaggct1260
ttcctgctcgccctcgagagtgccgtcgatcccgagtcgcactccgtgatcagtggcaag1320
gctcctgccggggccacgctgcggctgaagaagacgttcgccacgcccacctggtcgggc1380
acgatcaaggacaccctcgacaccacgatgaccgtcggcagcggcggcagctacacctgg1440
cacgtgaacccgtcgacccggccggtcgtcaaggcccgccagatcgaggtcatcggctcc1500
gagccgctgaagcggcagacctacacgggcacgaccgcgcccggacagccgacggagcag1560
gagttcgtcgtcgaccgggacgccgacgtcttcgaagcgaagctcgactgggccacgccc1620
gacgacctcgacctgtacgtcctgcgcaagaacgccgacggcagcctcacccaggtcggc1680
agttccgccggttccgtcggcgagaaggagcgggtcctcctcgacgacccggagcagggt1740
acgtacgtactccgcgtggagaactgggcttccgtcgcccccagttggaccctcaccgcg1800
tccctctacgacgccaccgtggacgagatcggcggcgtcatcgagaactggacgctctcc1860
tgcgagaaggacggaaaggtgcttcagcaggtgcccgtcgtcgtcgaccgtgggcagcgg1920
gtcaaggcggacttgaagaactgcgcgaagggctga1956
<210>21
<211>623
<212>prt
<213>actinoplanessp.
<400>21
metlysleuleumetilealametprotrpglnglyleuaspthrpro
151015
serseralaleuglyvalleuglyprocysvalarglysasnalaala
202530
asptrpthrvalaspgluleutyralaasnleuargtrpalaglutyr
354045
leumetarggluserasnglyservalthrcysgluasptyrglyasn
505560
ilealaaspglnvalphehisglyvalglyasptrpvalphethrpro
65707580
alaleutyraspvalaspsertyrglnvalaspglutyralalysphe
859095
leugluglnargaspmetaspprothrleuprovalglumethislys
100105110
tyralaargglypheileargaspleualaalagluilealaalaasp
115120125
proproaspvalvalglyphethrserthrphemetglnasnvalpro
130135140
serleualaleualaarggluleulyslysleualaproglyilearg
145150155160
thrvalleuglyglyserasncysaspglyalaglnglyproalaleu
165170175
hisargasnphegluglnleuaspphevalileserglygluglyglu
180185190
argalaleuproalaleuleuasnargileileargglygluserleu
195200205
alaaspvalproglyleusertrpargglyaspaspglyhisproval
210215220
valasnproproalathralaalaleuprophealametvalproala
225230235240
proglytyraspsertyrpheglnalaleugluargserprovalarg
245250255
hishisvalargprometleuvalleugluthrserargglycystrp
260265270
trpglyglualahisglncysthrphecysglyleuasnglyserasn
275280285
ileasppheargserlysalaprogluargilealaglngluvalarg
290295300
gluleualagluarghisglnileleuaspleuvalmetvalaspasn
305310315320
ileleuaspmetlystyrleuasnthralametprogluilealaala
325330335
leuaspcysaspleuargilehistyrgluilelysserasnmetasn
340345350
arggluglnleuserargleulysglualaasnvalleuphevalgln
355360365
proglyilegluserleuserserhisvalleuargleumetasplys
370375380
glyvalseralaalahisasnvalargmetleuargaspglyglnasp
385390395400
leuglyleuasnvalthrtrpserileleutyrglypheproglyglu
405410415
thrgluaspasptyrargglyleuleulyslysleualathrleuglu
420425430
hisleugluproprothrglyalatrpargilealaleugluargphe
435440445
serprotyrphegluaspprothrglnglyphemetpheargargpro
450455460
sergluiletyrasppheiletyrglnileproglnaspglnleutyr
465470475480
aspmetvalphephepheaspthrservalargglyileserglypro
485490495
ilegluaspglumetlysglnalacysgluglutrpalalysalatyr
500505510
proglnglythrleusertyrtrpthraspaspargglyargvalval
515520525
ilegluaspargargalasertrpprothrgluvalilegluleuasp
530535540
gluvalargserasnvaltyrleuglymetpheglncysalaalaarg
545550555560
gluglyileargargargleualaaspserglyhisvalvalglyglu
565570575
alagluleugluglumetleuargtyrphevalaspargglyleuala
580585590
phegluaspgluglyargtyrvalservalalaleuglyvalasppro
595600605
tyrargarglysleuvalglyglylysgluvalalaalaserleu
610615620
<210>22
<211>362
<212>prt
<213>actinoplanessp.
<400>22
metasnvalargphealagluargserthrleuargaspmetargala
151015
tyrargasplysgluserserasnalagluglyserserargphethr
202530
pheaspleuserserasngluleuvalleuproproleuprothrval
354045
leualaglyileglulysglyleuproargleualaargtyrproasp
505560
prothralaargaspleuthrgluaspilealaglyhisleucysval
65707580
serproaspgluvalalavalglyproglyseralaglyvalleugln
859095
glnileleuleualaleucysglylysglyaspgluvalvalhisgly
100105110
trpproglypheaspalatyrproleuleuvalalaileserglyala
115120125
thrglyvalhisvalproleuthralaserglyglyhisaspleuasp
130135140
gluileargthrargvalasnalaargthrargvalvalileleucys
145150155160
serprohisasnprothrglythrvalileaspglnaspgluleuhis
165170175
glypheleuargserleuproalahisvalvalalavalleuaspglu
180185190
alatyrvalglupheaspargglyalaasnproproglyleuproval
195200205
leuleusergluhisserasnthrvalvalleuargthrpheserlys
210215220
alatyrglyleualaglyleuargvalglytyralaalaglyproarg
225230235240
glnvalmetalathrvalarglysthralailepropheglyvalthr
245250255
argphealagluglnalaalametleuserleuargsergluaspglu
260265270
leucysgluargleualaalavalalaalaalaargglugluleuthr
275280285
alagluleuarggluleuargleuprovalleuleuserargalaasn
290295300
phevaltrpleuproleualaseralaalagluserphealaargthr
305310315320
alaalathralaglyvallysvalargalapheproglyhisglyval
325330335
argileservalglyglualaglualahisargthrleuleualaala
340345350
leuglyargalaaspargglyasntrpphe
355360
<210>23
<211>318
<212>prt
<213>actinoplanessp.
<400>23
metthrgluglnglyglyproserilealavalvalglyalaglygly
151015
valglyglytyrpheglyglyleuleualaalaalaglyhisaspval
202530
argpheleualaargglygluasnleualaalaleuargargglngly
354045
leuargilethrasnglyserseraspleuargvalproaspvalarg
505560
alaseralaaspprolysaspileglygluvalaspphevalleuleu
65707580
cysvallysthrserglnleuproalaalaleuaspalaleuglypro
859095
leuvalglygluhisthralavalvalthrvalglnasnglyvalglu
100105110
alaprogluglnvalalaalaargileglyargglyargvalleupro
115120125
glyservalargvalvalalaserthralaglyproglygluleuarg
130135140
hisvalglyproproglyalaleualaphethrglutrpaspserthr
145150155160
valseraspargvalalaargleuarggluvalleuargalaalaser
165170175
valservalprogluproseraspiletrpalaglyleutrpalalys
180185190
pheleuleuvalvalproileglyserleuglyalaalathrglygly
195200205
alathrileglygluleuargserargthrglythrargasnileleu
210215220
ilealaglymetarggluiletyrgluthrglyilelysleuglyile
225230235240
alaleuproalaalaalavalaspthralathrgluleumetaspgln
245250255
glnserproaspvalthrserserleuglnargaspileleualagly
260265270
argprosergluleuglualatrpthrglyalavalvalargleuala
275280285
argglyalaglyleuthralaprovalhisglumetleutyrgluleu
290295300
leualathrarggluserargthralaargserleuglnala
305310315
<210>24
<211>404
<212>prt
<213>actinoplanessp.
<400>24
metthraspvalileprothrgluphephethrgluproglyserasn
151015
prohisalathralaalaglutyrargserlyscysprovalhisarg
202530
ileasnvalproproglyalaaspalatyralavalleuglyasnlys
354045
valvalgluglualaleuglyaspserargleuserlysglnvalglu
505560
asnleuproalaargtyrargasplysalavalalaserserleuleu
65707580
valvalglyasnleuglyphealaaspalaprolyshisthrargleu
859095
lyslysproileserargalapheleuproalathrvalalaglnleu
100105110
argproargileglnaspilevalaspaspleuileaspthrphepro
115120125
gluasnglygluileaspleuleuserserphealaleuprometpro
130135140
leuthrvalilecysglutyrleuglyileprovalalaaspargpro
145150155160
leupheleuglutrpsertyrileleuserglnaspproleuglnhis
165170175
aspglualagluleulysalaalaserglugluphethrasptyrphe
180185190
thrlysleuvalalagluargargthraspleuargaspaspleuleu
195200205
sergluileileargalaargaspalaglyvaltyrsergluthrglu
210215220
leuleuserthrleuleuleuleuileilealaglyhislysthrval
225230235240
alaasnmetileglyasnglythralaleuleuleuarghisprogln
245250255
glnleuglumetleuargalathrprogluleuileproseralaile
260265270
glugluileleuargtyrgluglyseralaalatrpalaserleuarg
275280285
valalaalagluaspmetglnleualaglyvalaspileprolysgly
290295300
serphevalhisleuserleuserseralaglyargaspproaspval
305310315320
tyraspaspproaspglypheaspvalthrargserproasnarghis
325330335
leuserpheglyhisglyprohisphecysileglyalaproleugly
340345350
argleuglnglygluilealapheserthrleuleuargargleupro
355360365
argphegluleualavalproproglugluvalalatrpleuserasp
370375380
serserleuserargglyleuglualaleuproileargvalglyglu
385390395400
argleuproarg
<210>25
<211>100
<212>prt
<213>actinoplanessp.
<400>25
metproaspvallysleuproalaalaphehisvalleuthrglygly
151015
argargglnleuprovalgluglyalaasnilearggluvalleuval
202530
glyleuaspglnthrcysproglyvalleugluargleumetaspgln
354045
gluglyservallysargtyrvalasnvaltyrargasnaspserasp
505560
ileargserleuaspglyleugluthrlysvalgluhishisaspval
65707580
iletrpilevalproalavalalaglyglyserglualaalaargala
859095
glugluserarg
100
<210>26
<211>238
<212>prt
<213>actinoplanessp.
<400>26
metalaglyvallysaspalaglntyrvalthralaalathraspasp
151015
glyleuglyglythralaaspseralaalaleuleuaspaspleupro
202530
valthrvalargphegluilegluprovalargargpheleuserser
354045
alaleuglyglutyrglnlyscysleuaspserargaspalaaspgly
505560
valproserhisleuproargalaserglyleuleupheglyglnval
65707580
glyglyalagluilevalileseraspvalgluphevalproasnval
859095
argaspseraspgluservalmetalaglupheglualathrileala
100105110
proglnpheglyaspvaltyrlysasnproglyargglyphetrpser
115120125
aspgluglnglyvalleuglnalaileargglnglnseralaasngly
130135140
leugluleuleuglyserilehisserhisproasntrphisgluile
145150155160
glyproprohisgluargargglnargleusergluhisprothrgln
165170175
metaspglutyrleupheargglnsercystrpprovalasnvalile
180185190
trptyrvalhisgluserserglyglyilealahisargvalalaala
195200205
trpargproglyalagluglncysaspargleuaspileargilepro
210215220
alaalailehisgluglnphegluvalleuleuglugluglu
225230235
<210>27
<211>496
<212>prt
<213>actinoplanessp.
<400>27
metsertyrasnglythrserproargproproserileseralathr
151015
metthrleuilealathrglyvalservalleusertyralaleumet
202530
glnthrmetvalvalproalaleuhisvalleuglnvalglnleuhis
354045
thralaserthrtrpseralatrpileleuservalpheleuleuthr
505560
seralaalaserthrproleuleuserargleuglyaspargtyrser
65707580
lysarglysvalleuleuleuvalleuthrthrtyrleuileglythr
859095
valglycysalavalalaglyasnileglyvalleuilealacysarg
100105110
alavalglnglyvalserleualaalaileproleuserpheglyile
115120125
leuargaspvalleuprogluglnargleuargserglyleuglyleu
130135140
valserglythrileglyvalglyalaglyileglyleuvalvalgly
145150155160
glyleuvalvalasphisglnsertrpargtrpleuphealavalala
165170175
alavalleuileleuglyalaileglyleuvalalalystyrvalpro
180185190
aspglnargglyglualaglygluprovalaspvalproglyalaval
195200205
leuleualaleuvalleuvalalaleuleuleualaleuthrlysgly
210215220
thrsertrpglytrpalaserthrglythrleualaleupheglyala
225230235240
seralavalleuleuglyleuleuvalvalvalgluarglysserpro
245250255
alaproleuileaspproalavalvalalaglyargserphevalser
260265270
valhisglyalaalaphevalpheglyvalvalserphevalphetyr
275280285
valleuleuprothrtyralaglnthralaalaaspglnargleupro
290295300
glyglyglythrileglytyrglyleuglyalaaspvalthrmetala
305310315320
glyleuleuleuleuproglyserleuvalleuleuproalaglypro
325330335
leualaglyleuleuglnargleuthrservalargalathrleuala
340345350
serglyphealavalmetalavalglyalaileserleutrpalatrp
355360365
asnalaasnglytrpglnvalalavalglytyrleuvalvalglyleu
370375380
glyserglyleuvalleuserglyleuproservalileseraspleu
385390395400
thrglualaargargthralathralaasnglyvalasnthrvalval
405410415
argthralaglyglyvalvalglyserglnleualavalalaleuleu
420425430
alaalatrphisileserglyseraspthrproalaargaspglyphe
435440445
thrthralaphetrpilealaalaalavalalaalaalaglyglyleu
450455460
leucystrpvalglyilelysthrserthrleuargglyproargmet
465470475480
proglyvalthraspleuproargglnseralaglyglyvalargpro
485490495
<210>28
<211>313
<212>prt
<213>actinoplanessp.
<400>28
metgluleuaspleuarghisleuargtyrphevalalavalalaglu
151015
gluglyglyphethrargalaalaalaargleuhismetthrglnpro
202530
proleuservalalaileargglnleugluarggluleuglyleugln
354045
leuleuaspargthrglyasnargvalgluleuthrservalglyarg
505560
asppheleuthrhisalaargasnleuleuglnglntrpglnvalthr
65707580
valgluargmetargglnalaglyserglnaspvalgluargleuval
859095
valalapheargproalavalserargproleualahisargthrile
100105110
gluleuileargglulyshisproglutyrglnvalvalproargtyr
115120125
valprotrpthrgluglnthralacysleuglualaglyaspalaasp
130135140
valserphevalleugluproalaasptyrvalglyleugluargala
145150155160
thrvalalaleuleuproargvalvalcysleuproseralahisglu
165170175
leualaserargaspservalserileaspaspleusergluvalpro
180185190
ileileargprothrglyglyserproglutrpseraspphetrpgly
195200205
glygluvalcysproglylysargthrtrplysgluproprothrala
210215220
thrargleuaspglualaileaspleuvalalaleugluasnalaala
225230235240
alaleuvalprovalservalmetalavalglnhisargglnaspval
245250255
valpheileprovalthraspvalproalaalaargleuserleuala
260265270
trparggluglyseraspsergluleuvalargleualavalargcys
275280285
alaglnalaalaalaglnaspproalavalargthrleupheglyglu
290295300
proargprothrglythralaproala
305310
<210>29
<211>300
<212>prt
<213>actinoplanessp.
<400>29
metargargtrpalaalaglnserglyprogluaspleutyrpheval
151015
serasnleuhisalametthrthrlyshisaspprogluargleugln
202530
gluleuthrasphisglnleualaleuleuilealaalaglyvalpro
354045
glngluargleuphevalglnseraspleuileglngluhismetala
505560
leuthrtrpleuleuglucysthrcysthrpheglyglualaargarg
65707580
metvalglnphelysglulysserglnglyserasnservalargleu
859095
glyleuleuthrtyrprovalleumetalaalaaspileleuleuhis
100105110
glyalasergluvalprovalglyhisaspglnasnglnhisvalglu
115120125
leualaargthrleualaargargpheasnthrasptyrglygluval
130135140
phethrvalproglnalavalleuprovalalaalaalaargvalarg
145150155160
aspleualaalaprothrarglysmetserlysserserseraspgly
165170175
serglyilevaltyrvalleuaspserproglualavalargarglys
180185190
pheglnargalavalthraspglygluasnthrvalargtyralapro
195200205
aspgluglnproglyvalalaasnleuleugluileargalaalacys
210215220
thraspthrleuproseraspalaalalysglyileaspsertyrarg
225230235240
aspleulysglualaalaalaglualavalileserleuilealapro
245250255
valarggluargalaleuglnleuleuglugluargsergluleuala
260265270
lysileargalagluglyalaaspargalaargalaargserargasp
275280285
argleuaspargalaleuserleualaglyleulys
290295300
<210>30
<211>264
<212>prt
<213>actinoplanessp.
<400>30
metalaaspaspproleuvalileglyglythrsertyrserserarg
151015
leuilemetglythrglyglyalaproserleuaspvalleugluarg
202530
serleuvalalaserglythrgluleuthrthrvalalametargarg
354045
valaspproservallysglyservalleuservalleuaspargleu
505560
glyileglnvalleuproasnthralaglycysphethralaglyglu
65707580
alavalleuthralaargleualaargglualaleuglythraspleu
859095
vallysleugluvalilealaaspgluargthrleuleuproasppro
100105110
ilegluthrleuglualaalagluthrleuvalaspaspglyphethr
115120125
valleuprotyrthrasnaspaspprovalleualaarglysleugln
130135140
aspvalglycysalaalailemetproleuglyserproileglyser
145150155160
glyleuglyileargasnprohisasnpheglnleuilevalgluhis
165170175
alacysvalprovalileleuaspalaglyalaglythralaserasp
180185190
alaalaleualametgluleuglycysalaalavalmetleualaser
195200205
alavalthrargalaglngluprovalleumetalagluglymetarg
210215220
hisalavalglualaglyargleualahisargalaglyargilepro
225230235240
argarghisphealaglualaserserprothrgluglymetalaarg
245250255
leuaspprogluargproalaphe
260
<210>31
<211>392
<212>prt
<213>actinoplanessp.
<400>31
metserleuproproleuvalgluproalaalagluleuthrvalasp
151015
gluvalargargtyrserarghisleuileileproaspvalglymet
202530
aspglyglnlysargleulysasnalalysvalleucysvalglyala
354045
glyglyleuglyserproalaleumettyrleualaalaalaglyval
505560
glythrleuglyilevalglupheaspgluvalaspgluserasnleu
65707580
glnargglnileilehisserglnalaaspileglyargserlysala
859095
gluseralalysaspservalleuglyileasnprotyrvalasnval
100105110
ileleuhisglugluargleuglualagluasnvalmetaspilephe
115120125
serglntyraspleuilevalaspglythraspasnphealathrarg
130135140
tyrleuvalasnaspalacysvalleuleuasnlysprotyrvaltrp
145150155160
glyseriletyrargpheaspglyglnalaservalphetrpserglu
165170175
hisglyprocystyrargcysleutyrprogluproproproprogly
180185190
metvalprosercysalagluglyglyvalleuglyvalleucysala
195200205
serileglyserileglnvalasnglualailelysleuleualagly
210215220
ileglyaspproleuvalglyargleumetiletyraspalaleuglu
225230235240
metglntyrargglnvallysvalarglysaspproasncysalaval
245250255
cysglygluasnprothrvalthrgluleuileasptyrglualaphe
260265270
cysglyvalvalsergluglualaglnglualaalaleuglyserthr
275280285
ilethrprolysglnleulysglutrpileaspaspglygluasnile
290295300
aspileileaspvalarggluglnasnglutyrgluilevalserile
305310315320
proglyalaargleuileprolysasnglupheleumetglyglyala
325330335
leuglnaspleuproglnasplyslysilevalleuhiscyslysthr
340345350
glyvalargseralagluvalleualavalleulysseralaglyphe
355360365
alaaspalavalhisvalglyglyglyvalileglytrpvalasngln
370375380
ilegluproserlysprovaltyr
385390
<210>32
<211>424
<212>prt
<213>actinoplanessp.
<400>32
metsermetgluasnproargglyargarghisglyleuargleuala
151015
alaalaalavalalathralaserleumetserglyvalleuleuser
202530
thralaaspgluservalalaalaproproserthrtyrgluvallys
354045
glyvalaspthrserhishisasnhisaspalathrglylysproile
505560
asptrplysargvalalaglnserasnserphealapheleulysala
65707580
thrglnglythrglytyrlysaspprotrpphealaargaspphelys
859095
aspalaserglythrserleuleuargalaprotyrhisphepheasp
100105110
prolysserthrthraspglyglyalaglnalaasphispheilearg
115120125
alaalaargseralaglytyrthrglylysargalaglygluleupro
130135140
provalleuaspvalgluglythrtrpvalasnglylysgluvalcys
145150155160
prolysalaleuargalaaspglnleuthralapheleuasnargval
165170175
gluglualaphelysvalthrproilevaltyrthrargalaserphe
180185190
valasnglycysmetalaglylysglyglnvalphelysasphispro
195200205
leutrpleualaargtyrgluserglyserlysgluproglnaspval
210215220
proglyalaglyalatrpserleutrpglntyrthrglusergluala
225230235240
ileproglyleuproglylysasnglyalaalaglylysglyasparg
245250255
asnvaltyrargglyserleuaspglnleuargalaleualalysgly
260265270
glyglyalaproglnproglnproglythrsertrpprothrvallys
275280285
alaglyasplysglyvalaspvalalathrvalglnleuleuleugly
290295300
alahisglytyralathrthralaaspglyvalpheglythrglythr
305310315320
alaalalysvalglnalapheglnlysalagluglyleualaalaasp
325330335
glymetvalglyproalathrtrpalalysleuilealathrvallys
340345350
serglyserlysglythraspvalthralaleuglnargglnleuala
355360365
aspasnglytyraspvalthralaaspglyvalpheglyproalathr
370375380
thrserlysleuthralapheglnlysalalysglyleuthrserasp
385390395400
glyilealaglyproalathrtrpalalysleuvalserglyglythr
405410415
alaglyalathralathrthrser
420
<210>33
<211>331
<212>prt
<213>actinoplanessp.
<400>33
metglyvalgluargaspgluprothrargseralaargarggluleu
151015
alaleuleuleuargsertrptrpglualahisproasplysilethr
202530
glnglualaleualaargargilethrgluargglyvalargileser
354045
glnglumetleuserargtyrleuasnargserargprothrthrala
505560
argproaspvalileargthrmethisgluvalleuargargalapro
65707580
glugluleuaspvalalaleugluleuhisalaargalathralapro
859095
glnthrproproalagluglyalaalathrserglnproalaglyasp
100105110
alaglythralaalaprolysglyvalgluprothrseralaalapro
115120125
leuleuthrargthrprohisthrproargproalaserarglyslys
130135140
trpprotrpilealavalvalalaalaalavalvalglyalasergly
145150155160
leuthralaphemetthrleuglyaspglnargglnasnthrproarg
165170175
glyhisglyalathrproseralaserprothralaleuvalserpro
180185190
thralaglnglyserproalaglythrhisproproalaglucysarg
195200205
aspglusercyspheglyileaspalalystyralailecysglnasp
210215220
aspalaalathrtyrtyrthrglyargalahisglyvalleuvalglu
225230235240
leuargpheserproalacysglnalaalatrpalalysmetsergly
245250255
thrserglnglyaspvalvalargvalthrasnasnalaglyargser
260265270
arghistyrthrglnglntrpglyargaspalahisthrthrmetval
275280285
glualavalserproaspaspalalysalacysalaargthrproarg
290295300
glygluvalcysalathrlysalavalalaseralaproargaspala
305310315320
alaproglygluargalaalaproglyglyarg
325330
<210>34
<211>346
<212>prt
<213>actinoplanessp.
<400>34
metproalaargthrthrargthralahisthrthrargthrglyarg
151015
leualavalvalalaleualaalaleuthrcysalaglyleuvalthr
202530
glythralaalathralathrthrproaspserleuprothralalys
354045
argalaalaalaproaspalaalaalavalsertrpprothrleulys
505560
alaglyalaargglythrgluvalthralaleuglnhisleuleuile
65707580
alaargglyglnservalalavalaspglyglupheglyproalathr
859095
thrthralavallysalapheglnlysalaaspglyleuthralaasp
100105110
glyilevalglyproalathrtrpalalysleuvalprothrleuarg
115120125
glnglyalaglnglyalaalavallysalaalaglnthrleuleulys
130135140
thrargglyglnservalalavalaspglyglupheglyseralathr
145150155160
thrseralavallysalapheglnlysalalysglyleuseralaasp
165170175
glyvalvalglythrglnsertrpseralaleuleuthrseraspser
180185190
glyalaproserglyasnargalaalaphealaglnglnileleuasn
195200205
thrserglyilegluleualathrvalhisproglyglythrhisala
210215220
glyserthralaargglnasnileileaspthralaasnglylysgly
225230235240
alaleuthrserprotrpserasplysproasnglnargvalalaleu
245250255
aspthrargmetleuasnglyleuleulysleuleuserglnaspgly
260265270
tyrargileservalsergluilevalglyglyasphisserthrasn
275280285
serarghistyralaglyleuglypheaspileasntyrileasngly
290295300
arghisvalglygluseralaprohisglnglyleumetalaalacys
305310315320
arglysleuglyalathrgluvalleuglyproglyaspalaglyhis
325330335
serarghisvalhiscysglytrpproarg
340345
<210>35
<211>240
<212>prt
<213>actinoplanessp.
<400>35
metleuaspileaspgluleulysalaargaspseraspgluglyarg
151015
valproalaglyglyargproalathrgluthrleuthrleuglyleu
202530
aspargalagluleuprovalalathrgluleualaalaleuleuhis
354045
argvalprovalalaglyvalargleuprogluproalaasppheser
505560
alaleuproserhisvalilevalargileilealaleuileargglu
65707580
cysserserileglythrargvalthrtrpserleuthrleuglyala
859095
gluglnleuaspleuvalproargleuasphisleuproalaproasp
100105110
serilethrvalleugluthrglyhisproservalglyglutrparg
115120125
serserserasnpheglyleuleutyrphearglysglyprolysphe
130135140
leuservalvalaspglnargprogluserserarggluileileval
145150155160
aspaspprothrglnmetalavalpheleuleuglyleugluglycys
165170175
alatrpalagluvalthrargasnserglnphealaalaalaalaarg
180185190
aspleuvalasnlysglyleuvalmetargvalglyasphiscysval
195200205
thrleuprovalhismetargsertrpproleuglyalaalaleuleu
210215220
glyglythrleualaalaalaglylyslysseraspglyalathrglu
225230235240
<210>36
<211>846
<212>prt
<213>actinoplanessp.
<400>36
metseralavalleuglyvalleuleualaileserleualathrala
151015
proalahisalaalavalargseralaalaalavalaspvalcysarg
202530
seralaalaleuserlysalaargvalserthrtrpvalargleuglu
354045
hisargaspglythrtyrserargileargsergluleuservalglu
505560
valprogluasptrpproleualalysaspleuleuleusergluasp
65707580
serargargtyrvalalaalametsercysleuthrargthrasparg
859095
glyargglnargargtrpserglutrpargserserargprothrval
100105110
alaserthrlysserglyglyvallysvalvalaspargthrhisser
115120125
trpvalasnvaltyrargalahisileaspvalglythrtrpargval
130135140
argalaglyalagluargtrpthrvalglnleuglnalaproserala
145150155160
leuasnalaalaargtrpaspgluileargvalgluproglyalapro
165170175
glyalagluseralathrproargproaspgluglyargglyalathr
180185190
alaleuvaltrphisproglnasnhisargglulysalaalaalapro
195200205
alavalservalalaleulysprosertrpglnargsertrpalaala
210215220
glnasnaspargleuvalalavalalaleuaspargglyglytrpleu
225230235240
leutrpaspalathrseralaalaleuleuleutyralathrvalleu
245250255
tyrargargargseralaproprothrglnalaglngluargthrleu
260265270
argasnleuserleutrpalalysalaleuvalvalleuvalalaleu
275280285
thrsermetaspaspvalleuileargtyrvalglnargargglyasp
290295300
glyleuleuleuaspgluglnileproargglyasnalaphealaleu
305310315320
alaalavalilevalleuphecysvalglyargproargargargile
325330335
trpalaalaalaalavalleualavalprothrvalalaalaleupro
340345350
glntrpphegluleuserproglnargphevalseraspaspglutrp
355360365
alavalthrleualaalaglnglyvalalaalacyscysmetleuala
370375380
leuleuglyleuglyphevalthralaalatrpargleualavalasp
385390395400
glyaspleuleuprometserargarghisproglyhisalaargval
405410415
leuargleuargilealaglyprovalileleuvalcysthralaala
420425430
valalailecysphealaleualaglngluargasntrpglnargala
435440445
thrtrpleuseraspargseraspproalatyralathrglyglntrp
450455460
seraspargvaltrpglualavaltrpservalalaasnglyglnasp
465470475480
trpleusertrpglnalatrpleuleuthrglyvalalavalleuala
485490495
valleuargthrtrpargalaproalaservalserproleuaspasp
500505510
proalaaspargleuleupheleualaphephealailevalalaala
515520525
alaserglyglytyrpheleuglyasngluvalleuthrglyleutrp
530535540
ileproleusermetleualaleutyrtrpvalvalvalprophethr
545550555560
hisargservalleualaglnprophegluargserglyargproleu
565570575
alaaspseralaglyproglyalaargthrvalleuleualalysala
580585590
argsertyrarggluthrhisalagluleuargargleuaspglngly
595600605
leupheglyaspvalproprolysargseraspleugluglngluleu
610615620
seraspleuhisasntrpprothralaglyglyseraspargleupro
625630635640
alalysvalservalvalaspglyalaleualaleuglyproargasp
645650655
thrtrptrpalaasnglyserargcysalaargleualaleuvalpro
660665670
alavalproalaalaleuleuleualatrpvaltrplysvallysgly
675680685
glualatrphisalathrleuhisgluglnpheglyleuproaspval
690695700
leuleuleuphevalglyglumetvalmetphethrserseralaphe
705710715720
valleuglyalaleutrparghisleuproglyglnargglyalaala
725730735
lysalaleuprovalthrleualaphealaleuproileglyleuasp
740745750
alaleuvaltyrargphethrglygluserthralaasnleualaleu
755760765
alavalseralametleuphevalleuthrvalthrserilealaleu
770775780
asppheaspthrpheargglygluargargtyrtrpglnserargleu
785790795800
glyleuleuleuseriletyrglnmetargtyrtyrserleuglnala
805810815
alatyrleuilealaglnvalvalalametilethriletrpgluphe
820825830
phealagluproaspvalvalprolysproseraspserlys
835840845
<210>37
<211>548
<212>prt
<213>actinoplanessp.
<400>37
metleuthrleuthrvalproalametilethrpheglyvalpheleu
151015
ilealametvalmetileglyvalmetthrglnlysgluthralathr
202530
phealaaspphethrvalglyglyargargleuthralaprometala
354045
alaleuseralaglyalaseraspmetserglytrpleupheleugly
505560
leuproglyalavaltyrmetalaglyileglyalathrtrpileala
65707580
valglyleuilevalglythrtyrleuasntrpargphevalalapro
859095
argleuargthrtyrthrgluleualaglyasnservalthrleupro
100105110
sertyrleuglugluargphegluaspargserargmetleuargleu
115120125
leuseralailevalthrvalleuphephethrvaltyrvalalaser
130135140
glyleuvalalaglyglyleuleupheasngluilepheglyalaasp
145150155160
pheglupheglyleuthrvalphealavalvalilevalalatyrthr
165170175
ileleuglyglypheargalavalserilethrhisserileglngly
180185190
thrleumetpheleualaalaleuvalleuproalaleuglyleutrp
195200205
argleuglyglypheglyalaleuhisaspalaleuserasplysthr
210215220
proalaleuleuaspprovalalaglualaserphealaglyasnthr
225230235240
trpseralaglygluproleuglyalailealametileserleuleu
245250255
alatrpglyleuglytyrpheglyglnprohisileleuileargphe
260265270
metglyileargserthrlysaspileproleualaargargleugly
275280285
valglytrpvalvalvalvalleuglyglyserserleuileglyleu
290295300
alaglyilealavalleuaspgluproleuaspasnprogluthrval
305310315320
tyrilegluleuserthrhisleuvalasnprotrpilealaglyile
325330335
leuleuvalalavalleualaalailelysserthrvalaspsergln
340345350
leuleuvalseralathrserleuthrgluaspphetyrargalaphe
355360365
leuasnargargalaseraspthrleuleuleumetvalglyargleu
370375380
servalvalalavalalaleuvalalatyralailealaleusergly
385390395400
glyalavalleuaspilevalalatyralatrpalaglypheglyala
405410415
alapheglyprovalileileleuserleuphetrpproargmetthr
420425430
alaalaglyalametalaglymetvalthrglyalaleuthrvalphe
435440445
leutrplystyrileaspproleuleuglyproleugluserglyval
450455460
tyrglumetvalproglyvalleualaalathralaalaalaleuleu
465470475480
pheglylystyrvalglyargproproalaargalatrpileglyser
485490495
metgluhisalathrthrgluleuprothrprogluhisserthrthr
500505510
gluhisprothrmetgluglnglyserthrasptyrglythrhisgln
515520525
glyserthrasptyrglythrtyrglnglyleuproglnserglnasp
530535540
glntrpargpro
545
<210>38
<211>79
<212>prt
<213>actinoplanessp.
<400>38
metvalargargthrglyglyargargglyaspasnglyalaglnglu
151015
prothrserhisglyaspasphisalathrglyasnglyhisthrser
202530
thrargserserasncysthralaglnprolyscysthralahisthr
354045
asnprovalasnargseralathrthrgluglyhisalaglyalaala
505560
alaserproglyilealaglnglyprotrpglyglualavalgly
657075
<210>39
<211>651
<212>prt
<213>actinoplanessp.
<400>39
metthrvalaspthrprothrargalaalalysglulysleualagly
151015
leuglyleuaspleuthrgluhisalaglyhisglyphevalgluval
202530
valleuhisserproalaaspalaleualaleuglnvalglyglyphe
354045
sertrplysvalargvalproaspleuvalglnarggluseraspval
505560
asnalaalaasnargalatyralaalaalathrglythrserproleu
65707580
proserglyargaspsertyrargargleualaasptyrasnaspasp
859095
leuglyargmetalaaspglnasnproglyleuvalarglysphethr
100105110
leulyshislysserleugluglylysprovalhisglyvalgluile
115120125
thrhisaspvalthralavalaspaspglyargprovalpheleumet
130135140
metglyleuhishisalaargglutrpproserglygluhisalaile
145150155160
gluphealahisaspleuvalargasntyrglyseraspgluargile
165170175
thrserleuleuglnlysalaargvalleuvalvalprovalvalasn
180185190
valaspglypheglulysservalasnaspglyglnleuileaspleu
195200205
arggluileaspaspglyglythrglyserileleualathrprogly
210215220
asnalatyrlysarglysasncysargilevalaspglyleuserpro
225230235240
valalaglyglucysalaleualaserserproglyglypheglyala
245250255
glyvalaspleuasnargasntyrglyglyphetrpglyglyprogly
260265270
alaalaalagluservalglnalathrtyrargglyalaalaprophe
275280285
sergluprogluthrglnasnilearggluleuvalserserarggln
290295300
valthrglyleuilethrasnhisthrpheserasnleuvalleuarg
305310315320
proasnglyvalalaproaspthrvalglyproaspglyglnproile
325330335
glyasnproproaspglualaalaleulysgluleuglyaspargmet
340345350
alagluglnasnglytyrthrserglnhissertrpgluleutyrasp
355360365
thrthrglythrthrgluasptrpsertyrasnalathrglyglytyr
370375380
glytyrthrphegluileglyprohisgluphehisproprophepro
385390395400
gluvalvalaspglutyrvalglyalaglyglutyralaglylysgly
405410415
asnargglualapheleuleualaleugluseralavalaspproglu
420425430
serhisservalileserglylysalaproalaglyalathrleuarg
435440445
leulyslysthrphealathrprothrtrpserglythrilelysasp
450455460
thrleuaspthrthrmetthrvalglyserglyglysertyrthrtrp
465470475480
hisvalasnproserthrargprovalvallysalaargglnileglu
485490495
valileglysergluproleulysargglnthrtyrthrglythrthr
500505510
alaproglyglnprothrgluglngluphevalvalaspargaspala
515520525
aspvalpheglualalysleuasptrpalathrproaspaspleuasp
530535540
leutyrvalleuarglysasnalaaspglyserleuthrglnvalgly
545550555560
serseralaglyservalglyglulysgluargvalleuleuaspasp
565570575
progluglnglythrtyrvalleuargvalgluasntrpalaserval
580585590
alaprosertrpthrleuthralaserleutyraspalathrvalasp
595600605
gluileglyglyvalilegluasntrpthrleusercysglulysasp
610615620
glylysvalleuglnglnvalprovalvalvalaspargglyglnarg
625630635640
vallysalaaspleulysasncysalalysgly
645650
- 还没有人留言评论。精彩留言会获得点赞!