一种金属硫蛋白基因MT20、其编码得到的金属硫蛋白及其表达和应用的制作方法
本发明属于基因工程技术领域,具体涉及一种金属硫蛋白基因、其编码得到的金属硫蛋白及其表达和应用。
背景技术:
金属硫蛋白(metallothionein,简称mt)是一类普遍存在于生物体内的金属结合蛋白。金属硫蛋白是具有结合金属能力和高诱导特性的低分子量蛋白质。富含半胱氨酸的短肽,对多种重金属有高度亲和性。它是分子质量较低,半胱氨酸残基和金属含量极高的蛋白质。与其结合的金属主要是镉、铜和锌,广泛地存在于从微生物到人类各种生物中,其结构高度保守。
重金属生物修复是利用生物材料进行重金属治理的技术,而重金属抗性基因是生物修复的重要资源。基于基因改造的生物材料可以突破自然生物材料的性状限制,获得生物量、生长速度和修复效率的优化组合,来自微生物的重金属抗性基因已经在高效修复生物材料的开发方面显示出巨大的潜力。早在1998年,naturebiotechnology(rughetal.,1998)就报道了稳定表达细菌汞还原基因mera的黄杨(yellowpoplar,又译作鹅掌楸),该转基因植物展示了一种生态友好的植物修复途径。来自沼泽红假单胞菌(rhodopseudomonaspalustris)的砷甲基化基因arsm最近被用于了构建基因工程菌,该工程菌展示了高效修复砷污染土壤的潜力(chenetal.,2014)。少数研究也展示了基因改造生物材料在镉污染修复上的潜力。例如,shim等人将酵母的镉抗性基因ycf1转入杨树,促进了该植株在镉污染土壤中的生长,也增强了该植株的镉富集能力(shimetal.,2013);kang等人将三个微生物镉抗性基因sppcs、gshi和mnta转入大肠杆菌,使转基因菌株的镉富集能力提高了25倍(kangetal.,2007)。
现阶段对于重金属耐受/富集效果的金属硫蛋白来源往往来自于动物、植物,对微生物来源的金属硫蛋白研究较少。
技术实现要素:
本发明的目的在于获得一种重金属耐受/富集效果的金属硫蛋白基因,从而得到由其编码的金属硫蛋白,进而将其应用于重金属污染治理领域中。
本发明第一方面是提供一种,金属硫蛋白基因,其碱基序列如seqidno:1所示,由该基因编码得到的氨基酸序列如seqidno:2所示。本发明通过以下步骤分离得到上述金属硫蛋白基因:
s1.用铜溶液污染土壤,避光恒温25度培养14天
s2.使用商用dna提取试剂盒提取土壤全基因组
s3.使用二代illumia测序平台hiseq对基因组进行全测序
s4.通过通用生物信息学方法对测序序列进行质检、过滤、组装
s5.从公开基因数据库uniprot和genbank中搜索所有有关的金属硫蛋白基因序列
s6.手动检查所述金属硫蛋白基因序列可靠性,编号后将可靠金属硫蛋白基因序列汇总,经软件翻译形成金属硫蛋白氨基酸序列数据库,所述金属硫蛋白氨基酸序列数据库见seqidno:3~seqidno:54;
s7.利用生物信息学检索方法blast,在测序组装的土壤基因组中搜索可能的金属硫蛋白基因序列,以所述金属硫蛋白氨基酸序列数据库作为检索模板
s8.获得的所述可能的金属硫蛋白基因序列进行手动检查,核对序列的核苷酸偏好性、金属结合位点等信息,进一步确定高可靠性的金属硫蛋白基因序列
s9.功能验证
9.1将所述高可靠性的金属硫蛋白基因序列通过化学合成获得高可靠性疑似金属硫蛋白序列,所述高可靠性疑似金属硫蛋白序列直接连接至pet28a(+)质粒,然后转化大肠杆菌bl21(de3),得到转化菌株;
9.2通过dropassay试验测定所述大肠杆菌bl21(de3)及所述转化菌株的镉抗性,使用所述大肠杆菌bl21(de3)作为对照,当最小抑制浓度下该转化菌株成活,认为所述高可靠性的金属硫蛋白基因序列为有功能/活性的金属硫蛋白基因;
9.3通过测定生长管内600nm处吸光值获得所述大肠杆菌bl21(de3)的生长曲线;
9.4收获生长曲线测定中的所述大肠杆菌bl21(de3);
9.5通过强酸消解所述大肠杆菌bl21(de3),使用原子吸收测定溶液镉浓度并计算所述大肠杆菌bl21(de3)内镉吸收量。
本发明的优点和有益效果为:
本发明公开的金属硫蛋白基因,来源于土壤微生物,该金属硫蛋白基因重金属耐受性好,在1.0m浓度镉溶液中仍然可以存活生长。并且对镉金属富集效应明显,因而在重金属污染治理等行业具有较大的应用潜力。
附图说明
图1是本发明公开的大肠杆菌在不同浓度镉溶液中的生长情况。
图2是本发明公开的大肠杆菌生长曲线。
对于本领域普通技术人员来讲,在不付出创造性劳动的前提下,可以根据以上附图获得其他的相关附图。
具体实施方式
为了使本技术领域的人员更好地理解本发明方案,下面结合具体实施例进一步说明本发明的技术方案。
实施例一
金属硫蛋白基因筛选获得的过程:
s1.用铜溶液污染土壤,避光恒温25度培养14天;
s2.使用商用dna提取试剂盒(dneasypowermaxsoilkit)按照其说明书规定流程进行操作提取土壤全基因组,凝胶电泳测定dna质量;
s3.提交商业测序,使用二代illumia测序平台hiseq对基因组进行全测序
s4.通过通用生物信息学方法对测序序列进行质检、过滤、组装
s5.从公开基因数据库uniprot和genbank中搜索所有有关的金属硫蛋白基因序列
s6.手动检查所述金属硫蛋白基因序列可靠性,编号后将可靠金属硫蛋白基因序列汇总,经过软件翻译形成金属硫蛋白氨基酸序列数据库,所述金属硫蛋白氨基酸序列数据库见seqidno:3~seqidno:54;
s7.利用生物信息学检索方法blast,在测序组装的土壤基因组中搜索可能的金属硫蛋白基因序列,以所述金属硫蛋白氨基酸序列数据库作为检索模板,
s8.获得的所述可能的金属硫蛋白基因序列进行手动检查,核对序列的核苷酸偏好性、金属结合位点等信息,进一步确定高可靠性的金属硫蛋白基因序列,所述可靠性的金属硫蛋白基因序列碱基序列如seqidno:1所示,编号为mt20。
实施例二
9.1将所述高可靠性的金属硫蛋白基因序列mt20通过化学合成获得高可靠性疑似金属硫蛋白序列如seqidno:2所示(通过商业公司genewiz,suzhou,完成),所述高可靠性疑似金属硫蛋白序列直接连接至pet28a(+)质粒,质粒通过电转化转入宿主大肠杆菌escherichiacoli(e.coli)bl21(de3),得到转化菌株;
实施例三
功能验证的详细过程。
9.2最小抑制浓度(mic),通过测量不同cd浓度培养基中细菌细胞的生长能力来评估cd的mic值。首先,含有转化菌株的mt20及其相应的空白对照(空pet28a(+))在lauriabertani(lb)-琼脂平板上于29℃生长过夜。然后,将含有细菌菌落的重组质粒扩散到含有各种浓度(0-16mm)cd(包括卡那霉素(50mg/l))和诱导剂(iptg-isopropyl-b-d-1-thiogalactopyranoside,100mg/l)的lb-plate平板中。)测定他们的mic。
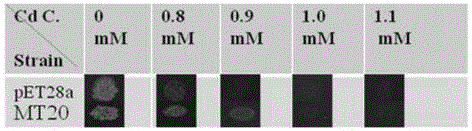
测定空白对照大肠杆菌escherichiacoli(e.coli)bl21(de3),cd最小抑制浓度,dropassay试验测定所述大肠杆菌bl21(de3);见图1,在cd浓度为0.8mmol/l时,空白对照大肠杆菌escherichiacoli(e.coli)bl21(de3),图中标号为pet28a无法存活,其最小抑制浓度为cd0.8mmol/l;而转化菌株,图中标号为mt20,在cd浓度为0.8mmol/l、0.9mmol/l时均可存活,其最小抑制浓度为cd1.0mmol/l证明该转化菌株镉耐受性好。
9.3用于筛选不同cd溶液中更耐受菌株的生长模式(生长曲线)。
对于该测定,首先,将储存的细菌细胞转移到含有卡那霉素和iptg的lb-琼脂平板中,并在37℃下孵育过夜以进行活化。然后将单个细菌菌落转移到含有抗生素卡那霉素(50μg/ml),iptg(100μg/ml)和1.0mmcd的100mllb液体培养基中。将细菌样品在37℃,160-180rpm连续摇动。摇动12-14小时后,通过nanodrop分光光度计每隔1小时检查细菌样品在600nm处的吸光值(光密度od600),持续24小时。将所有读数绘制成图2,以获得1.0mmcd中每种细菌的生长曲线。
如图2所示,其中pet1~pet3均为空白对照大肠杆菌escherichiacoli(e.coli)bl21(de3),转化菌株图中标号为mt20,在生长过程中,转化菌株的吸光值(菌群数量)均高于三个空白对照大肠杆菌样品,并且维持较高增长,在第13个小时达到最高峰,转换菌株的菌群数量比空白对照大肠杆菌的菌群数量高约50%,证明其在cd环境下耐受性能好,寿命长。
9.4、9.5cd结合试验
检查od600后,使用高速离心机收集细菌细胞。将收集的生物质在65℃温育24-48小时以进行干燥。然后将干燥的生物质加入
sequencelisting
<110>中国科学院遗传与发育生物学研究所农业资源研究中心
<120>一种金属硫蛋白基因mt20、其编码得到的金属硫蛋白及其表达和应用
<130>1
<160>54
<170>patentinversion3.5
<210>1
<211>261
<212>dna
<213>unknown
<220>
<223>unknown
<400>1
atggccgaatgcgcgtggtgccacaacgagtacgacaaaacgttttctgtgacgtacgga60
gatcagtcgtacgtctttgattgcttcgaatgcgcgatcacgatgctggcgccaacgtgc120
gctcactgtggctgtcgcatcatcggacacggcgatgaggtggacgggcgcttctactgc180
tgcgcttcctgtgtaagggccgccgaaccggtggatctgcacgaccgcgcgactgctggc240
cacgtgggtggcacacgctag261
<210>2
<211>86
<212>prt
<213>unknown
<220>
<223>unknown
<400>2
metalaglucysalatrpcyshisasnglutyrasplysthrpheser
151015
valthrtyrglyaspglnsertyrvalpheaspcyspheglucysala
202530
ilethrmetleualaprothrcysalahiscysglycysargileile
354045
glyhisglyaspgluvalaspglyargphetyrcyscysalasercys
505560
valargalaalagluprovalaspleuhisaspargalathralagly
65707580
hisvalglyglythrarg
85
<210>3
<211>57
<212>prt
<213>thermosynechococcusvulcanus
<400>3
metthrthrvalthrglnmetlyscysalacysprohiscysleucys
151015
ilevalserleuasnaspalailemetvalaspglylysprotyrcys
202530
sergluvalcysalaasnglythrcyslysgluasnserglycysgly
354045
hisalaglycysglycysglyserala
5055
<210>4
<211>54
<212>prt
<213>microcystisaeruginosa
<400>4
metilealavalthrthrmetlyscysalacysglysercysthrcys
151015
glnvalserilealaaspalailelyslysasnaspglntyrtyrcys
202530
cysglnalacysalaasnglyhisvallysglulysglycysglyhis
354045
proglycysvalcysgly
50
<210>5
<211>57
<212>prt
<213>thermosynechococcussp.
<400>5
metthrthrvalthrglnmetlyscysalacysprohiscysleucys
151015
ilevalserleuseraspalailemetvalaspglylysprotyrcys
202530
sergluvalcysalaasnglythrcyslysgluserasnglycysgly
354045
hisserglycysglycysglyserala
5055
<210>6
<211>57
<212>prt
<213>methylobacteriumradiotolerans
<400>6
metalaservalaspvalglumetvallyscysalacysglnaspcys
151015
valcysvalileprovalalalysalavalserargaspglylysala
202530
tyrcyscysaspaspcysalaaspglyhislysasphisalaglycys
354045
gluhisalaglycysalacyshisgly
5055
<210>7
<211>56
<212>prt
<213>microcystisaeruginosa
<400>7
metilealavalthrmetmetlyscysalacysglusercysleucys
151015
valvalleuilealaaspalailelyslysasnaspglntyrtyrcys
202530
serglnalacysalaasnglyhisvalasngluasnglulysglycys
354045
glyhisglnglycysglycysval
5055
<210>8
<211>56
<212>prt
<213>microcystisaeruginosa
<400>8
metilealavalthrmetmetlyscysalacyslysprocysleucys
151015
valvalserilealaaspalailelysgluasnasplystyrtyrcys
202530
serglnalacysalaasnglyhisvalasngluasnglulysglycys
354045
glyhisglnglycysglycysval
5055
<210>9
<211>56
<212>prt
<213>synechococcus
<400>9
metthrserthrthrleuvallyscysalacysgluprocysleucys
151015
asnvalaspproserlysalaileaspargasnglyleutyrtyrcys
202530
serglualacysalaaspglyhisthrglyglyserlysglycysgly
354045
histhrglycysasncyshisgly
5055
<210>10
<211>53
<212>prt
<213>mycobacteriumtuberculosis
<400>10
metargvalileargmetthrasntyrglualaglythrleuleuthr
151015
cysserhisgluglycysglycysargvalargilegluvalprocys
202530
hiscysalaglyalaglyaspalatyrargcysthrcysglyaspglu
354045
leualaprovallys
50
<210>11
<211>48
<212>prt
<213>mycobacteriumcaprae
<400>11
metthrasntyrglualaglythrleuleuthrcysserhisglugly
151015
cysglycysargvalargilegluvalprocyshiscysalaglyala
202530
glyaspalatyrargcysthrcysglyaspgluleualaprovallys
354045
<210>12
<211>53
<212>prt
<213>mycobacteriumbovis
<400>12
metargvalileargmetthrasntyrglualaglythrleuleuthr
151015
cysserhisgluglycysglycysargvalargilegluvalprocys
202530
hiscysalaglyalaglyaspalatyrargcysthrcysglyaspglu
354045
leualaprovallys
50
<210>13
<211>55
<212>prt
<213>leptolyngbyasp.
<400>13
metalathrvalthrglnmetlyscysalacysgluprocysleucys
151015
ilevalaspileserlysalaileglnlysaspglyglntyrtyrcys
202530
sergluglycysalaserglyhisglyaspasnserlysglycysgly
354045
histhrglycysasncyshis
5055
<210>14
<211>54
<212>prt
<213>filamentouscyanobacterium
<400>14
metalathrvalthrglnmetlyscysalacysglusercysleucys
151015
valvalaspileserlysalaileglulysgluglyglntyrtyrcys
202530
glyglualacysalaasnglyhissergluglyserthrglycysgly
354045
hisproglycysasncys
50
<210>15
<211>54
<212>prt
<213>halomicronemahongdechloris
<400>15
metthrthrvalthrglnmetlyscysalacysaspsercysleucys
151015
ilevalaspthrserlysalavalglulysgluglyhistyrtyrcys
202530
serglualacysalaasnglyhisprogluglyserglycysglyhis
354045
thrglycysthrcyshis
50
<210>16
<211>55
<212>prt
<213>geitlerinemasp.
<400>16
metthrthrvalthrglnmetlyscysalacysglusercysleucys
151015
valvalaspthrasplysalavalglulysaspglyglntyrtyrcys
202530
serglualacysalaasnglyhisproaspglyserglycysglyhis
354045
glnglycysthrcyshisala
5055
<210>17
<211>55
<212>prt
<213>phormidiumtenue
<400>17
metthrthrvalthrglnmetlyscysalacysaspsercysleucys
151015
valvalaspthrserglnalavalglulysaspglyhistyrphecys
202530
serglualacysalaasnglyhisprogluglyseralaglycysgly
354045
hisproglycysglycysasn
5055
<210>18
<211>54
<212>prt
<213>lyngbyaconfervoides
<400>18
metthrthrvalthrglnmetlyscysalacysaspsercysleucys
151015
ilevalasnthrserlysalavalglulysgluglyhistyrtyrcys
202530
seraspalacysalaasnglyhisprogluglyserglycysglyhis
354045
thrglycysthrcyshis
50
<210>19
<211>54
<212>prt
<213>kamptonema
<400>19
metthrthrvalthrglnmetlyscysalacyssersercysleucys
151015
valvalserleuthrglualaileglulysasnglyglntyrtyrcys
202530
serasnalacysalaaspglyhisproasnglythrglycysglyhis
354045
alaglycysglycyshis
50
<210>20
<211>48
<212>prt
<213>spirulinasubsalsa
<400>20
vallyscysalacysserthrcysglucysmetvalserproasplys
151015
alaileglulysaspglylystyrtyrcysglyglualacysalaasn
202530
glyhisthraspglyserhisglycysglyhisproglycysasncys
354045
<210>21
<211>54
<212>prt
<213>acaryochlorismarina
<400>21
metalathrvalthrglnmetlyscysalacysglusercysleucys
151015
ilevalthrileseraspalaileglnlysglyglyglntyrphecys
202530
glyglnalacysalaaspglyhisproserglyserglycysglyhis
354045
thrglycysglycyshis
50
<210>22
<211>54
<212>prt
<213>acaryochlorismarina
<400>22
metalathrvalthrglnmetlyscysalacysglusercysleucys
151015
ilevalthrileseraspalaileglnlysglyglyglntyrphecys
202530
glyglnalacysalaaspglyhisproserglyserglycysglyhis
354045
thrglycysglycyshis
50
<210>23
<211>53
<212>prt
<213>tolypothrixcampylonemoides
<400>23
metthrasnvalthrglnleulyscysalacysgluprocysleucys
151015
valvalserleugluaspalaileglnlysaspglylystyrtyrcys
202530
serglualacysalagluglyhisglnthrmetglnglycysglyhis
354045
serglycysglycys
50
<210>24
<211>53
<212>prt
<213>tolypothrixcampylonemoides
<400>24
metthrasnvalthrglnleulyscysalacysgluprocysleucys
151015
valvalserleugluaspalaileglnlysaspglylystyrtyrcys
202530
serglualacysalagluglyhisglnthrmetglnglycysglyhis
354045
serglycysglycys
50
<210>25
<211>53
<212>prt
<213>calothrixsp.
<400>25
metthrthrvalthrmetmetlyscysalacysgluargcysleucys
151015
valvalserthralaaspalaileglulysgluglylystyrtyrcys
202530
serglnalacysalaaspglyhislysaspglulysglycysalahis
354045
serglycysglycys
50
<210>26
<211>52
<212>prt
<213>cyanobacteriumaponinum
<400>26
thrthrvalthrglnmetlyscysalacysprosercysleucysile
151015
ileaspileserglnalaileserargaspglyhistyrtyrcysser
202530
thralacysalagluglyhislysgluglygluglycysglyhisser
354045
glycysglycys
50
<210>27
<211>53
<212>prt
<213>scytonematolypothrichoides
<400>27
metthrservalthrglnmetlyscysalacysgluprocysleucys
151015
ilevalserleugluasnalaileglnlysaspglulystyrtyrcys
202530
serglualacysalagluglyhislysthrmetlysglycysglyhis
354045
asnglycysglycys
50
<210>28
<211>53
<212>prt
<213>limnoraphisrobusta
<400>28
metthrservalthrglnmetlyscysalacysglusercysleucys
151015
valvalserleugluseralailelyslysaspglylysprotyrcys
202530
serglualacysalaasnglyhisserasnglyserglycysglyhis
354045
thrglycysthrcys
50
<210>29
<211>54
<212>prt
<213>oscillatoriabrevis
<400>29
metthrthrvalthrglnilelyscysalacysprosercysleucys
151015
valvalserleuthrglualaileglulysserglylyssertyrcys
202530
serseralacysalaaspglyhisproasnglythrglycysglyhis
354045
thrglycysglucyshis
50
<210>30
<211>53
<212>prt
<213>fischerella
<400>30
metthrthrvalthrglnmetlyscysalacysglusercysleucys
151015
ilevalserilegluaspalaileglnlysaspasnlystyrtyrcys
202530
serglualacysalaaspglyhisglnthrthrlysglycysglyhis
354045
serglycysglycys
50
<210>31
<211>54
<212>prt
<213>leptolyngbyaohadii
<400>31
metthrthrvalthrglnmetlyscysalacysseraspcysleucys
151015
ilevalasnleuasnaspalailemetlysaspglylysalatyrcys
202530
glyaspalacysalaasnglyhisthrglyglyserglycysglyhis
354045
thrglycysglycyshis
50
<210>32
<211>53
<212>prt
<213>acaryochlorismarina
<400>32
metalathrvalthrglnmetlyscysalacysglusercysleucys
151015
ilevalthrileseraspalaileglnlysglyglyglntyrphecys
202530
glyglnalacysalaaspglyhisproserglyserglycysglyhis
354045
thrglycysargcys
50
<210>33
<211>54
<212>prt
<213>phormidesmispriestleyi
<400>33
metthralavalthrglnmetlyscysalacysgluprocysleucys
151015
ilevalthrthrgluglyalavalglnlysaspglylysleutyrcys
202530
sergluvalcysalaaspglyhisproasnglyhisglyaspcysgly
354045
hislysglycysthrcys
50
<210>34
<211>54
<212>prt
<213>microcoleusvaginatus
<400>34
metthrthralathrglnthrlyscysalacysprosercyssercys
151015
valvalasnvalserglualaileglulysaspglylysthrtyrcys
202530
serseralacysalaaspglyhisproasnglyserglycysglyhis
354045
thrglycysglucyshis
50
<210>35
<211>54
<212>prt
<213>geminocystisherdmanii
<400>35
thrvalthrglnmetlyscysalacysprosercysleucysileval
151015
aspilealaseralaileglnlysaspasnglntyrphecysserasp
202530
alacysalaasnglyhislysgluglythrthrglycysserhisser
354045
glycysglycyshisgly
50
<210>36
<211>54
<212>prt
<213>geitlerinemasp.
<400>36
metthrthrvalthrglnmetlyscysalacysglusercysleucys
151015
valvalasnleuseraspalavalhislysaspglulystyrtyrcys
202530
cysglualacysalaasnglyhisglnserglyaspglycysglyhis
354045
serglycysglycyshis
50
<210>37
<211>53
<212>prt
<213>mastigocladopsisrepens
<400>37
metthrservalthrglnmetlyscysalacysgluprocysleucys
151015
ilevalserleugluaspalaileglnlysaspasplystyrtyrcys
202530
serglualacysalagluglyhisglnthrmetlysglycysglyhis
354045
asnglycysglycys
50
<210>38
<211>52
<212>prt
<213>cyanothecesp.
<400>38
thrvalthrglnmetlyscysalacyssersercysvalcysileval
151015
aspleuseraspalaileglnlysaspglylystyrtyrcysserasp
202530
alacysalaasnglyhisproaspglyalaglycysserhishisgly
354045
cysglucyshis
50
<210>39
<211>53
<212>prt
<213>mastigocoleustestarum
<400>39
metalaaspvalthrsermetlyscysalacysalaaspcysleucys
151015
ilevalserleulysaspalailealalysasnglyglntyrtyrcys
202530
sergluvalcysalaasnglyhisvalaspglyserglycysglyhis
354045
thrglycyslyscys
50
<210>40
<211>53
<212>prt
<213>calothrixsp.
<400>40
metthrthrvalthrglnmetlyscysalacysglusercysleucys
151015
ilevalserleuserseralavalmetlysgluglylysprotyrcys
202530
glyglualacysalaasnglyhisalaaspglylysglycysglyhis
354045
thrglycysglucys
50
<210>41
<211>56
<212>prt
<213>pseudanabaenasp.
<400>41
metalaseralathrleuvallyscysalacysserlyscysleucys
151015
valileaspproseraspalaileglualaasnglylystyrtyrcys
202530
cyslysalacysalaserglyhisvalaspglythrasnaspserhis
354045
cysseraspvalglycysglucys
5055
<210>42
<211>58
<212>prt
<213>geminocystissp.
<400>42
metthrthralathrilethrglnmetlyscysalacysprosercys
151015
leucysilevalaspileglythralaleuglnlysgluglylystyr
202530
phecysserthralacysalagluglyhislysgluglythrthrgly
354045
cysserhisthrglycysglycysasngly
5055
<210>43
<211>53
<212>prt
<213>calothrixsp.
<400>43
metthrthrvalthrglnmetlyscysalacysglusercysleucys
151015
ilevalserleuserseralavalmetlysgluglylysprotyrcys
202530
glyglualacysalaasnglyhisglnaspglylysglycysglyhis
354045
thrglycysglycys
50
<210>44
<211>53
<212>prt
<213>rivulariasp.
<400>44
metalaalavalaspleumetlyscysalacysasplyscysleucys
151015
ilevallysvalgluthralaileaspargaspglylyshistyrcys
202530
serglualacysalagluglyhislysthrilethrglycysglyhis
354045
serglycysglycys
50
<210>45
<211>55
<212>prt
<213>coleofasciculuschthonoplastes
<400>45
metthrthralathrglnthrglncysalacysaspsercysalacys
151015
metvalserthraspseralavalglnlysaspglylystyrtyrcys
202530
seraspalacysalaasnglyhisproasnglyalaglycysglyhis
354045
serglycysglucyshisala
5055
<210>46
<211>53
<212>prt
<213>stanieriacyanosphaera
<400>46
metserthrvalthrsermetlyscysalacysaspargcysleucys
151015
valvalserleugluaspalavallyslysaspglylystyrtyrcys
202530
cysglualacysalaasnglyhisthraspglyserglycysglyhis
354045
glnglycysglycys
50
<210>47
<211>54
<212>prt
<213>hydrocoleumsp.
<400>47
metthrthrvalthrglnmetlyscysalacysglusercysleucys
151015
ilevalserilegluseralavallyslysasnglyglnasntyrcys
202530
serglualacysalaasnasnhisproaspglyalaglycysglyhis
354045
gluglycysglucysasn
50
<210>48
<211>53
<212>prt
<213>chamaesiphonminutus
<400>48
metserthrvalthrglnmetlyscysalacysglusercysleucys
151015
ilevalserleuseraspalailevallysaspglylyshistyrcys
202530
glyaspalacysalaasnglyhisproalaglyglnglycysglyhis
354045
thrglycysglycys
50
<210>49
<211>55
<212>prt
<213>tolypothrixbouteillei
<400>49
metthrthrvalserglnmetlyscysalacyslyssercysleucys
151015
valvalserleuseraspalaleumetlysaspglylysalatyrcys
202530
glyglualacysalaasnglyhisthrasnglyglucyscysglyhis
354045
thrglycysaspcyshisala
5055
<210>50
<211>54
<212>prt
<213>hapalosiphonaceae
<400>50
metthrthrvalthrglnmetlyscysalacysglusercysleucys
151015
valvalserleuthraspalavalilelysaspglylysprotyrcys
202530
glyglualacysalaasnglyhisproasnglygluglycysglyhis
354045
glnglycysglycyshis
50
<210>51
<211>53
<212>prt
<213>aliterellaatlantica
<400>51
metthrthralathrglnthrglncysalacysglusercyshiscys
151015
provalsergluthrglualavalglnlysaspglylysthrtyrcys
202530
serglualacysalaglnglyhisproaspglylysglycysglyhis
354045
alaglycysaspcys
50
<210>52
<211>53
<212>prt
<213>aliterellaatlantica
<400>52
metthrthralathrglnthrglncysalacysglusercyshiscys
151015
provalsergluthrglualavalglnlysaspglylysthrtyrcys
202530
serglualacysalaglnglyhisproaspglylysglycysglyhis
354045
alaglycysaspcys
50
<210>53
<211>53
<212>prt
<213>cyanobacteriumstanieri
<400>53
metthrthrvalthrglnmetlyscysalacysprosercysleucys
151015
ilevalasnleuseraspalaileglnlysasnasphistyrtyrcys
202530
cysglnalacysalaaspglyhisproasnglyserglycysglyhis
354045
thrglycysglycys
50
<210>54
<211>53
<212>prt
<213>xenococcussp.
<400>54
thrvalthrglnmetlyscysalacysprosercysleucysileval
151015
asnvalseraspalaileserlysgluglylystyrtyrcysserasp
202530
alacysalalysglyhissergluglyalaglycysserhisalagly
354045
cysglycyshisala
50
- 还没有人留言评论。精彩留言会获得点赞!