一种产软骨素的重组谷氨酸棒杆菌及其应用的制作方法
本发明涉及一种产软骨素的重组谷氨酸棒杆菌及其应用,属于生物工程技术领域。
背景技术
硫酸软骨素是一种重要的糖胺聚糖,由葡萄糖醛酸和n-乙酰半乳糖胺构成的二糖单位通过β1-3和β1-4糖苷键交替连接形成。硫酸软骨素广泛应用于医疗保健、化妆品和食品添加剂等领域。目前硫酸软骨素主要是以动物组织为原料通过化学提取法获得。由于不同动物组织中的硫酸软骨素硫酸化程度相差较大,很难获得纯度高且质量均一的产品。最近兴起的生物酶法催化代表了硫酸软骨素和生产方法的重要发展方向。该方法是分别以软骨素作为底物经过软骨素硫酸转移酶的硫酸化等修饰,获得高纯度,硫酸化程度均一的产品。但受限于软骨素生产量不足,质量不高,目前很难通过此方法工业化生产硫酸软骨素。因此获得高质量、高产量的软骨素是促进硫酸软骨素生产方法革新,提高硫酸软骨素产量和质量的重要节点。
在自然条件下,大肠杆菌k4能够分别产生果糖化修饰的软骨素构成其荚膜的重要成分。因此人们希望通过不断优化大肠杆菌k4代谢途径以提高果糖化的软骨素的产量。但是大肠杆菌k4对人和动物有致病性,会引起肠道感染,败血症等疾病,带有较大的安全隐患。此外,大肠杆菌k4生产是发生了果糖化修饰的软骨素,而不是软骨素,因此需要额外的脱果糖处理。第三,果糖化软骨素合成过程受到复杂的调控,造成获得的产量偏低。这些缺点不但会造成生产成本偏高,还会增加下游产物的加工、纯化难度。而在其他微生物细胞中如大肠杆菌bl21和枯草芽孢杆菌中重组构建的软骨素合成侧重对代谢途径进行优化,缺乏对软骨素合成关键酶的改造,造成途径效率不高,造成软骨素产量低。
因此有必要探索、构建安全高效的微生物细胞工厂,提高软骨素和前体的产量与质量。
技术实现要素:
为了解决上述问题,本发明对工业安全菌株谷氨酸棒杆菌(corynebacteriumglutamicum)进行改造,大幅提高了软骨素的产量。
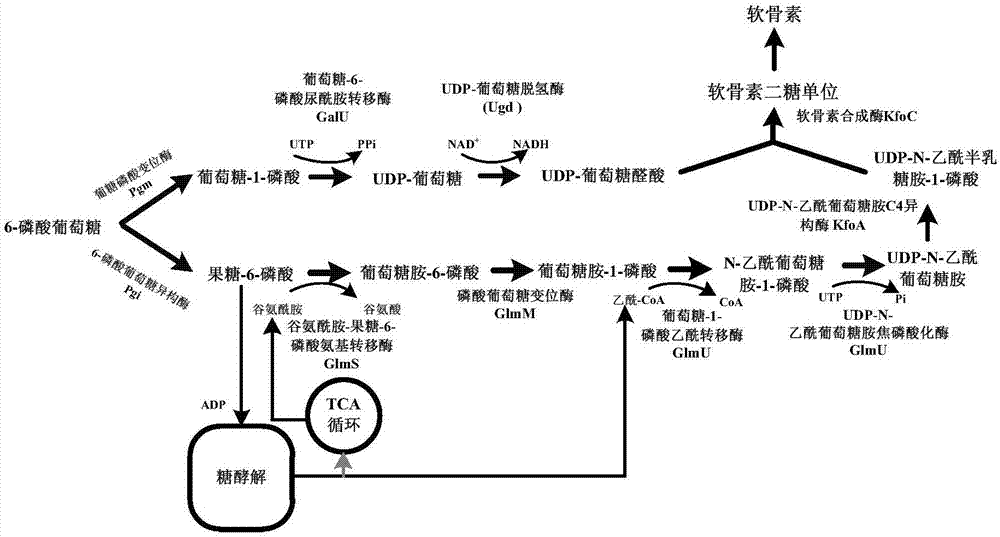
本发明提供了一种重组谷氨酸棒杆菌,所述谷氨酸棒杆菌异源整合表达了udp-n-乙酰葡萄糖胺c4异构酶kfoa和软骨素合成酶kfoc,并通过以质粒表达谷氨酰胺-果糖-6-磷酸氨基转移酶glms、磷酸葡萄糖变位酶glmm、udp-n-乙酰葡萄糖胺焦磷酸化酶/葡萄糖-1-磷酸乙酰转移酶双功能酶glmu和葡萄糖-6-磷酸尿酰胺转移酶galu基因,以加强软骨素合成底物udp-n-乙酰葡萄糖胺和udp-葡萄糖醛酸的积累,在谷氨酸棒杆菌中构建了软骨素的合成通路。
所述软骨素合成酶是野生型或者h357g/n363s双突变体。选择软骨素合成酶h357g/n363s双突变体,可以大幅提高谷氨酸棒杆菌生产软骨素的能力。
进一步的,将单个软骨素合成关键酶kfoc、kfoa和udp-葡萄糖脱氢酶ugd进行融合改造,形成人工酶复合体,增加了软骨素合成关键酶之间的关联性,进一步促进软骨素的积累。
所述的udp-n-乙酰葡萄糖胺c4异构酶kfoa和软骨素合成酶kfoc来源于大肠杆菌k4(escherichiacolio5:k4:h4(l):h4,escherichiacolik4)。kfoa的氨基酸序列如seqidno.1所示,kfoc的氨基酸序列如seqidno.2所示。所述的谷氨酰胺-果糖-6-磷酸氨基转移酶glms,磷酸葡萄糖变位酶glmm,udp-n-乙酰葡萄糖胺焦磷酸化酶/葡萄糖-1-磷酸乙酰转移酶双功能酶glmu,葡萄糖-6-磷酸尿酰胺转移酶galu,udp-葡萄糖脱氢酶ugd来源于谷氨酸棒杆菌corynebactriumglutamicumatcc13032。
在本发明的一种实施方式中,glmu、galu以pxmj19质粒为表达载体,以ppyc为启动子,形成组成型表达框,在谷氨酸棒杆菌中持续表达。glms和glmm以pec-xk99e质粒为表达载体,以ptrc启动子,形成诱导型表达框,在谷氨酸棒杆菌中iptg诱导表达。glms、glmm、glmu氨基酸序列分别如seqidno.3、seqidno.4、seqidno.5,galu的氨基酸序列包括seqidno.6和seqidno.7所示。
在本发明的一种实施方式中,采用ggsggsggggs蛋白linker融合将kfoc,kfoa和ugd串联融合形成人工酶复合体。复合体中kfoc,kfoa和ugd的摩尔比例为1:1:1。ugd的氨基酸序列包括seqidno.9、seqidno.10所示。
本发明的有益效果:本发明利用谷氨酸棒杆菌产软骨素,与其它软骨素生产菌相比,该发明具有非常大的优势。首先,本发明过程中使用的谷氨酸棒杆菌为安全菌株(generallyrecognizedassafe,gras),完全满足医疗卫生和食品安全生产的要求;与枯草芽孢杆菌、大肠杆菌相比谷氨酸棒杆菌培养成本低,发酵过程易于控制,且发酵菌体密度高,菌体无裂解,适应高密度发酵。本发明在打通软骨素在谷氨酸棒杆菌中合成通路的基础上,侧重对软骨素合成关键酶进行深入改造:通过对软骨素合成关键酶kfoc进行随机突变,提高酶活性,以及通过对kfoc,kfoa和ugd进行人工复合体构建提高多步代谢步骤之间的关联性,最终重组谷氨酸棒杆菌生产的软骨素的产量达到13.4g/l。本发明不但解决了大肠杆菌k4菌株生产软骨素携带的安全隐患,避免了软骨素的果糖化修饰,而且大幅提高了软骨素的产量,为通过重组谷氨酸棒杆菌工业化生产软骨素打下基础。
附图说明
图1:重组谷氨酸棒杆菌生产软骨素和前体的代谢途径及相关的酶类。
图2:kfoc的突变位点和kfoa、kfoc、ugd酶复合体的构建示意图;其中gslinker的氨基酸序列为ggsggsggggs,g为甘氨酸,s为丝氨酸。
图3:重组谷氨酸棒杆菌软骨素产量图;其中,c.glutamicumchon1:kfoa和kfoc野生型基因组整合表达,glmu和galu以pxmj19为载体表达,glms和glmm以pec-xk99e表达;c.glutamicumchon2:kfoa野生型和kfoc(h357g/n363s)突变体基因组整合表达,glmu和galu以pxmj19为载体表达,glms和glmm以pec-xk99e表达;c.glutamicumchon3:kfoa野生型和kfoc(h357g/n363s)突变体以及ugd串联形成人工酶复合体,并在基因组整合表达,glmu和galu以pxmj19为载体表达,glms和glmm以pec-xk99e表达。
具体实施方式
菌株:谷氨酸棒杆菌c.glutamicumatcc13032,质粒:pxmj19,pec-xk99e,pk18mobsacb。菌株构建培养基:bhis:脑心浸液肉汤37g/l,山梨糖醇90g/l;软骨素发酵培养基:葡萄糖:40g/l,玉米浆干粉:20g/l,(nh4)2so4:30g/l,kh2po4:1g/l,k2hpo4:1g/l,mgso4:25g/l,mops(3-吗啉丙磺酸):42g/l,谷氨酸棒杆菌培养温度为30℃。分子实验工具酶:pcr实验所用酶为primerstarmaxdnapolymerase(takara);限制性内切酶为fastdigest(thermoscientific);一步克隆实验所用酶为clonexpress(诺唯赞)。软骨素的产量测定:向装有1ml硼砂硫酸(500ml浓h2so4中溶解4.77g硼砂)的玻璃管中加入适当稀释的200μl样品,混匀后在沸水浴中煮15min,冷却至室温。加入50μl咔唑试剂(100ml无水乙醇中溶解0.125g咔唑),混匀后再煮沸15min。测定波长530nm处的吸光值。用不同浓度(l0、20、30、40、50μgml-1)d-葡萄糖醛酸标准品进行相同反应,绘制标准曲线。标准曲线方程:y=0.0149x-0.0239,r2=0.9991(x,样品中葡萄糖醛酸含量(μgml-1);y,吸光度a530)。软骨素产量计算公式:软骨素含量(g/l)=(标准曲线测出的浓度*稀释倍数*2.067)/1000。
实施例1:将ptrc-kfoa-kfoc整合到谷氨酸棒杆菌基因组
以ptrc-f与ptrc-r为引物,以pec-xk99e为模板扩增ptrc,以kfoa-f与kfoa-r为引物,以大肠杆菌k4基因组为模板扩增kfoa;以kfoc-f与kfoc-r为引物,以大肠杆菌k4基因组为模板扩增kfoc;将获得的ptrc、kfoa和kfoc以ptrc-f和kfoc-r为引物进行重叠延伸pcr,获得ptrc-kfoa-kfoc表达框。以glgaup-f和glgadown-r为引物,以ptrc-kfoa-kfoc和谷氨酸棒杆菌glga基因上、下游500bp同源臂(两个500bp同源臂分别通过引物glgaup-f与glgaup-r、glgadown-f与glgadown-r以谷氨酸棒杆菌atcc13032基因组为模板进行pcr获得)为模板再次进行重叠延伸pcr,获得h1-ptrc-kfoa-kfoc-h2。
将h1-ptrc-kfoa-kfoc-h2以一步克隆的方式连接入bamhi和hindiii双酶切的pk18mobsacb质粒,获得质粒pk18mobsacb-h1-ptrc-kfoa-kfoc-h2。将质粒pk18mobsacb-h1-ptrc-kfoa-kfoc-h2通过电穿孔(12.5kv/cm,5ms)方式转化谷氨酸棒杆菌。并在含有25μg/ml的bhis平板上筛选第一步重组的转化子。第一步重组的阳性转化子在bhis液体培养基中过夜培养后,涂布于含有20%蔗糖的bhis平板上进行蔗糖致死反选,获得第二步重组的转化子。以glga-check-f和glga-check-r为引物,通过菌落pcr验证成功整合ptrc-kfoa-kfoc的单克隆。
实施例2:重组菌c.glutamicumchon1的构建
corynebacteriumglutamicumatcc13032接种于5mlbhis液体培养基,在30℃200rpm培养24h,收集菌体,采用细胞基因组提取试剂盒提取基因组dna。第一步:pxmj19-glmu-galu的构建。设计引物ppyc-f/ppyc-r、glmu-f/glmu-r、galu-f/galu-r,以提取的c.glutamicumatcc13032基因组dna为模板,采用标准的pcr扩增体系和程序,扩增获取ppyc、glmu、galu基因。以扩增的glmu、galu为模板,以glmu-f和galu-r为引物进行重叠延伸pcr获得相连的glmu-galu片段。以pxmj19-f/pxmj19-r为引物扩增缺失ptac和laciq区域的线性pxmj19质粒。将glmu-galu片段和线性质粒pxmj19分别进行一步克隆拼接反应。拼接反应体系转化jm109感受态细胞。阳性转化子进行质粒测序,与序列比对,重组质粒pxmj19-glmu-galu构建成功。第二步:pec-xk99e-glms-glmm的构建。设计glms-f/glms-r、glmm-f/glmm-r,以提取的c.glutamicumatcc13032基因组dna为模板,采用标准的pcr扩增体系和程序,扩增获取glms、glmm基因。以扩增的glmu、galu为模板,以glmu-f和galu-r为引物进行重叠延伸pcr获得相连的glms-galm片段。将glmu-galu片段和由kpni和sali双酶切线性质粒pec-xk99e分别进行一步克隆拼接反应。拼接反应体系转化jm109感受态细胞。阳性转化子进行质粒测序,与序列比对,重组质粒pec-xk99e-glms-glmm构建成功。第三步:电转化质粒pxmj19-glmu-galu和pec-xk99e-glms-glmm进入谷氨酸棒杆菌,构建c.glutamicumchon1。质粒pxmj19-glmu-galu和pec-xk99e-glms-glmm通过电穿孔(12.5kv/cm,5ms)方式转化实施例1中构建的整合了ptrc-kfoa-kfoc的谷氨酸棒杆菌,在含有卡那霉素和氯霉素的bhis平板上筛选阳性转化子,c.glutamicumchon1构建成功。
表1ptrc-kfoa-kfoc整合到谷氨酸帮杆菌基因组以及重组质粒pxmj19-glmu-galu构建所用引物
实施例3:kfoc(h357g/n363s)突变体的构建与筛选
按照试剂盒的操作手册,以kfoc-f与kfoc-r为引物,利用diversifytmpcrrandommutagenesiskitpcr随机突变试剂盒对kfoc基因进行随机突变。将随机突变的pcr片段按照实施例1中的描述将ptrc启动子以及和kfoa基因组装形成ptrc-kfoa-kfoc(随机突变体)表达框,并按照实施例1中的描述,将ptrc-kfoa-kfoc(随机突变体)表达框整合到谷氨酸棒杆菌基因组。含有ptrc-kfoa-kfoc(随机突变体)的重组谷氨酸棒杆菌单菌落库在96孔板中进行发酵培养。待葡萄糖耗尽后(40小时后)用96孔板离心收集菌体,并用去离子水洗涤菌体2次。菌体进行适当倍数的稀释后,用硫酸咔唑方法测定软骨素的含量,并筛选出产量最高的突变体。产量最高的突变体经过kfoc基因测序,确定产软骨素产量最高的重组谷氨酸棒杆菌中kfoc发生突变的类型为h357g/n363s。
实施例4:c.glutamicumchon2的构建
按照实施例1、2和3中的表述,将pxmj19-glmu-galu和pec-xk99e-glms-glmm质粒分别电转化进入整合了kfoa野生型和kfoc(h357g/n363s)突变体的谷氨酸棒杆菌中,获得c.glutamicumchon2菌株。
实施例5:c.glutamicumchon3的构建:
第一步构建:ptrc-kfoa-linker-kfoc。该表达框的构建方式与实施例1中所述基本相同。不同之处是ptrc-kfoa-linker-kfoc中kfoa的终止密码子taa和kfoc的核糖体结合位点(rbs序列aggagg)被编码ggsggsggggslinker的核苷酸序列ggtggaagtggaggtagtggaggtggaggtagt所替代。替代方法为重叠延伸pcr。第二步构建:ptrc-kfoa-linker-kfoc-linker-ugd。在第一步构建的基础上,将谷氨酸棒杆菌udp-葡萄糖脱氢酶的编码基因ugd通过编码ggsggsggggslinker的核苷酸序列ggaggtagcggtggaagcggtggaggtggaagc与ptrc-kfoa-linker-kfoc融合形成ptrc-kfoa-linker-kfoc-linker-ugd。构建方法同样为重叠延伸pcr。为避免dna出现正向重复,连接kfoc和ugd的ggsggsggggslinker的核苷酸序列与第一步中连接kfoa与kfoc的核苷酸序列有所不同,但两个linker的氨基酸序列保持一致。将ptrc-kfoa-linker-kfoc-linker-ugd转化谷氨酸棒杆菌。第三步:电转化质粒pxmj19-glmu-galu和pec-xk99e-glms-glmm进入步骤(2)中已经转化了ptrc-kfoa-linker-kfoc-linker-ugd的谷氨酸棒杆菌,构建c.glutamicumchon3。
实施例6:重组谷氨酸棒杆菌摇瓶发酵生产软骨素实验
将构建的重组谷氨酸棒杆菌菌株:c.glutamicumchon1,c.glutamicumchon1和c.glutamicumchon3(三种菌株基因型见图3说明)的单克隆接于5mlbhi培养基,置于200rpm,30℃过夜培养。培养12-16小时后,按1%的接种量转接于250ml三角摇瓶(装液量25ml发酵培养基)中。置于200rpm,28℃培养,并添加1.5mmiptg进行诱导基因表达。待葡萄糖耗尽后终止发酵实验,并通过硫酸咔唑法测定软骨素含量。结果如图3所示,c.glutamicumchon3的软骨素产量可以达到6.8g/l。
实施例7:c.glutamicumchon3发酵罐发酵实验
第一步,种子液的制备:准备c.glutamicumchon3种子液。将c.glutamicumchon3的单克隆接入100ml发酵培养基(葡萄糖10g/l),置于200rpm,28℃过夜培养。第二步,补料发酵:在3l发酵罐中装入900ml发酵培养基,灭菌后接入第一步的种子液。发酵培养基中初始葡萄糖浓度为40g/l。控制发酵温度为28℃,搅拌转速为500rpm。之后每4小时左右取一次样品,测定菌体浓度和葡萄糖含量。ph用氨水控制在6.5-7.0之间,当葡萄糖浓度降低至5g/l以下时将葡萄糖浓度补至15g/l.当葡萄糖消耗速率明显下降时(低于2g/h),停止补料。待残留葡萄糖耗尽后结束发酵。
sequencelisting
<110>江南大学
<120>一种产软骨素的重组谷氨酸棒杆菌及其应用
<160>36
<170>patentinversion3.3
<210>1
<211>339
<212>prt
<213>escherichiacolik4
<400>1
metasnileleuvalthrglyglyalaglytyrileglyserhisthr
151015
serleucysleuleuasnlysglytyrasnvalvalileileaspasn
202530
leuileasnsersercysgluserileargargilegluleuileala
354045
lyslyslysvalthrphetyrgluleuasnileasnasnglulysglu
505560
valasnglnileleulyslyshislyspheaspcysilemethisphe
65707580
alaglyalalysservalalagluserleuilelysproilephetyr
859095
tyraspasnasnvalserglythrleuglnleuileasncysalaile
100105110
lysasnaspvalalaasnpheilepheserserseralathrvaltyr
115120125
glygluserlysilemetprovalthrgluaspcyshisileglygly
130135140
thrleuasnprotyrglythrserlystyrilesergluleumetile
145150155160
argaspilealalyslystyrseraspthrasnpheleucysleuarg
165170175
tyrpheasnprothrglyalahisgluserglymetileglygluser
180185190
proalaaspileproserasnleuvalprotyrileleuglnvalala
195200205
metglylysleuglulysleumetvalpheglyglyasptyrprothr
210215220
lysaspglythrglyvalargasptyrilehisvalmetaspleuala
225230235240
gluglyhisvalalaalaleusertyrleupheargaspasnasnthr
245250255
asntyrhisvalpheasnleuglythrglylysglytyrservalleu
260265270
gluleuvalserthrpheglulysileserglyvalargileprotyr
275280285
gluilevalserargargaspglyaspilealaglusertrpserser
290295300
proglulysalaasnlystyrleuasntrplysalalysarggluleu
305310315320
gluthrmetleugluaspalatrpargtrpglnmetlysasnproasn
325330335
glytyrile
<210>2
<211>686
<212>prt
<213>escherichiacolik4
<400>2
metserileleuasnglnalaileasnleutyrlysasnlysasntyr
151015
argglnalaleuserleupheglulysvalalagluiletyraspval
202530
sertrpvalglualaasnilelysleucysglnthralaleuasnleu
354045
serglugluvalasplysleuasnarglysalavalileaspileasp
505560
alaalathrlysilemetcysserasnalalysalaileserleuasn
65707580
gluvalglulysasngluileileserlystyrarggluilethrala
859095
lyslyssergluargalagluleulysgluvalgluproileproleu
100105110
asptrpproseraspleuthrleuproproleuprogluserthrasn
115120125
asptyrvaltrpalaglylysarglysgluleuaspasptyrproarg
130135140
lysglnleuileileaspglyleuserilevalileprothrtyrasn
145150155160
argalalysileleualailethrleualacysleucysasnglnlys
165170175
thriletyrasptyrgluvalilevalalaaspaspglyserlysglu
180185190
asnileglugluilevalarggluphegluserleuleuasnilelys
195200205
tyrvalargglnlysasptyrglytyrglnleucysalavalargasn
210215220
leuglyleuargalaalalystyrasntyrvalalaileleuaspcys
225230235240
aspmetalaproasnproleutrpvalglnsertyrmetgluleuleu
245250255
alavalaspaspasnvalalaleuileglyproarglystyrileasp
260265270
thrserlyshisthrtyrleuasppheleuserglnlysserleuile
275280285
asngluileprogluileilethrasnasnglnvalalaglylysval
290295300
gluglnasnlysservalasptrpargilegluhisphelysasnthr
305310315320
aspasnleuargleucysasnthrpropheargphepheserglygly
325330335
asnvalalaphealalyslystrpleupheargalaglytrppheasp
340345350
glugluphethrhistrpglyglygluaspasnglupheglytyrarg
355360365
leutyrarggluglycystyrpheargservalgluglyalametala
370375380
tyrhisglngluproproglylysgluasngluthraspargalaala
385390395400
glylysasnilethrvalglnleuleuglnglnlysvalprotyrphe
405410415
tyrarglyslysglulysilegluseralathrleulysargvalpro
420425430
leuvalseriletyrileproalatyrasncysserlystyrileval
435440445
argcysvalgluseralaleuasnglnthrilethraspleugluval
450455460
cysilecysaspaspglyserthraspaspthrleuargileleugln
465470475480
gluhistyralaasnhisproargvalargpheileserglnlysasn
485490495
lysglyileglyseralaserasnthralavalargleucysarggly
500505510
phetyrileglyglnleuaspseraspasppheleugluproaspala
515520525
valgluleucysleuaspgluphearglysaspleuserleualacys
530535540
valtyrthrthrasnargasnileasparggluglyasnleuileser
545550555560
asnglytyrasntrpproiletyrserargglulysleuthrserala
565570575
metilecyshishispheargmetphethralaargalatrpasnleu
580585590
thrgluglypheasngluserileserasnalavalasptyraspmet
595600605
tyrleulysleusergluvalglyprophelyshisileasnlysile
610615620
cystyrasnargvalleuhisglygluasnthrserilelyslysleu
625630635640
aspileglnlysgluasnhisphelysvalvalasngluserleuser
645650655
argleuglyilelyslystyrlystyrserproleuthrasnleuasn
660665670
glucysarglystyrthrtrpglulysilegluasnaspleu
675680685
<210>3
<211>564
<212>prt
<213>corynebacteriumglutamicumatcc13032
<400>3
leuaspalagluilealalysalaproleuproaspserileleugly
151015
ileglyhisthrargtrpalathrhisglyglyprothraspvalasn
202530
alahisprohisvalvalserasnglylysleualavalvalhisasn
354045
glyileilegluasnphealagluleuargsergluleuseralalys
505560
glytyrasnphevalseraspthraspthrgluvalalaalaserleu
65707580
leualagluiletyrasnthrglnalaasnglyaspleuthrleuala
859095
metglnleuthrglyglnargleugluglyalaphethrleuleuala
100105110
ilehisalaasphisaspaspargilevalalaalaargargasnser
115120125
proleuvalileglyvalglygluglygluasnpheleuglyserasp
130135140
valserglypheileasptyrthrarglysalavalgluleualaasn
145150155160
aspglnvalvalthrilethralaaspasptyralailethrasnphe
165170175
aspglyserglualavalglylyspropheaspvalglutrpaspala
180185190
alaalaalaglulysglyglypheglyserphemetglulysgluile
195200205
hisaspglnproalaalavalargaspthrleumetglyargleuasp
210215220
gluaspglylysleuvalleuaspgluleuargileaspglualaile
225230235240
leuargservalasplysilevalilevalalacysglythralaala
245250255
tyralaglyglnvalalaargtyralailegluhistrpcysargile
260265270
prothrgluvalgluleualahisglupheargtyrargaspproile
275280285
leuasnglulysthrleuvalvalalaleuserglnserglygluthr
290295300
metaspthrleumetalavalarghisalaarggluglnglyalalys
305310315320
valvalalailecysasnthrvalglyserthrleuproarggluala
325330335
aspalaserleutyrthrtyralaglyprogluilealavalalaser
340345350
thrlysalapheleualaglnilethralasertyrleuleuglyleu
355360365
tyrleualaglnleuargglyasnlysphealaaspgluvalserser
370375380
ileleuaspserleuargglumetproglulysileglnglnvalile
385390395400
aspalaglugluglnilelyslysleuglyglnaspmetalaaspala
405410415
lysservalleupheleuglyarghisvalglypheprovalalaleu
420425430
gluglyalaleulysleulysgluilealatyrleuhisalaglugly
435440445
phealaalaglygluleulyshisglyproilealaleuvalgluglu
450455460
glyglnproilephevalilevalproserproargglyargaspser
465470475480
leuhisserlysvalvalserasnileglngluileargalaarggly
485490495
alavalthrilevalilealaglugluglyaspglualavalasnasp
500505510
tyralaasnpheileileargileproglnalaprothrleumetgln
515520525
proleuleuserthrvalproleuglnilephealacysalavalala
530535540
thralalysglytyrasnvalaspglnproargasnleualalysser
545550555560
valthrvalglu
<210>4
<211>447
<212>prt
<213>corynebacteriumglutamicumatcc13032
<400>4
metthrargleupheglythraspglyvalargglyleualaasnglu
151015
valleuthralaproleualaleulysleuglyalaalaalaalahis
202530
valleuthralaglulysargvalaspglyargargprovalalaile
354045
valglyargaspproargvalserglyglumetleualaalaalaleu
505560
seralaglymetalaserglnglyvalaspvalileargvalglyval
65707580
ileprothrproalavalalapheleuthraspasptyrglyalaasp
859095
metglyvalmetileseralaserhisasnprometproaspasngly
100105110
ilelysphepheseralaglyglyhislysleuproasphisvalglu
115120125
aspgluilegluargvalmetaspserleuproalagluglyprothr
130135140
glyhisglyvalglyargvalilegluglualathraspalaglnasp
145150155160
argtyrleugluhisleulysglualavalprothrserleuglugly
165170175
ilelysilevalvalaspalaalaasnglyalaalaservalvalala
180185190
prothralatyrglualaalaglyalathrvalilealailehisasn
195200205
lysproaspsertyrasnileasnmetaspcysglyserthrhisile
210215220
aspglnvalglnalaalavalleulyshisglyalaaspleuglyleu
225230235240
alahisaspglyaspalaaspargcysleualavalasplysaspgly
245250255
asnleuvalaspglyaspglnilemetalaleuleualailealamet
260265270
lysgluasnglygluleuarglysasnthrleuvalglythrvalmet
275280285
serasnleuglyleulysilealametaspglualaglyilethrleu
290295300
argthrthrlysvalglyaspargtyrvalleugluaspleuasnala
305310315320
glyglypheserleuglyglygluglnserglyhisilevalleupro
325330335
asphisglythrthrglyaspglythrleuthrglyleuserilemet
340345350
alaargmetalagluthrglylysserleuglygluleualaglnala
355360365
metthrvalleuproglnvalleuileasnvalprovalserasplys
370375380
serthrilevalserhisproservalvalalaalailealagluala
385390395400
glualagluleuglyalathrglyargvalleuleuargalasergly
405410415
thrglugluleupheargvalmetvalglualaglyasplysglugln
420425430
alaargargilealaglyargleualaalavalvalalagluval
435440445
<210>5
<211>485
<212>prt
<213>corynebacteriumglutamicumatcc13032
<400>5
metseralaserasppheserseralavalvalvalleualaalagly
151015
alaglythrargmetlysseraspleuglnlysthrleuhisserile
202530
glyglyargserleuileserhisserleuhisalaalaalaglyleu
354045
asnprogluhisilevalalavalileglyhisglyargaspglnval
505560
glyproalavalalaglnvalalaglugluleuasparggluvalleu
65707580
ilealaileglnglugluglnasnglythrglyhisalavalglncys
859095
alametaspglnleugluglyphegluglythrileilevalthrasn
100105110
glyaspvalproleuleuthrasphisthrleuseralaleuleuasp
115120125
alahisvalgluvalprothralavalthrvalleuthrmetargleu
130135140
aspaspprothrglytyrglyargilevalargasnglugluglyglu
145150155160
valthralailevalgluglnlysaspalaseralagluvalglnala
165170175
ileaspgluvalasnserglyvalphealapheaspalaalaileleu
180185190
argseralaleualagluleulysseraspasnalaglnglygluleu
195200205
tyrleuthraspvalleuglyilealaargglygluglyhisproval
210215220
argalahisthralaalaaspalaarggluleualaglyvalasnasp
225230235240
argvalglnleualaglualaglyalagluleuasnargargthrval
245250255
ilealaalametargglyglyalathrilevalaspproalathrthr
260265270
trpileaspvalgluvalserileglyargaspvalileilehispro
275280285
glythrglnleulysglygluthrvalileglyaspargvalgluval
290295300
glyproaspthrthrleuthrasnmetthrileglyaspglyalaser
305310315320
valileargthrhisglypheaspserthrileglygluasnalathr
325330335
valglyprophethrtyrileargproglythrthrleuglyproglu
340345350
glylysleuglyglyphevalgluthrlyslysalathrileglyarg
355360365
glyserlysvalprohisleuthrtyrvalglyaspalathrilegly
370375380
glugluserasnileglyalaserservalphevalasntyraspgly
385390395400
gluasnlyshishisthrthrileglyserhisvalargthrglyser
405410415
aspthrmetpheilealaprovalthrvalglyaspglyalatyrser
420425430
glyalaglythrvalilelysaspaspvalproproglyalaleuala
435440445
valserglyglyargglnargasnilegluglytrpvalglnlyslys
450455460
argproglythralaalaalaglnalaalaglualaalaglnasnval
465470475480
hisasnglnglugly
485
<210>6
<211>315
<212>prt
<213>corynebacteriumglutamicumatcc13032
<400>6
metserleuproileaspgluhisvalasnalavallysthrvalval
151015
valproalaalaglyleuglythrargpheleuproalathrlysthr
202530
valprolysgluleuleuprovalvalaspthrproglyilegluleu
354045
ilealaalaglualaalagluleuglyalathrargleualaileile
505560
thralaproasnlysalaglyvalleualahisphegluargserser
65707580
gluleuglugluthrleumetgluargglylysthraspglnvalglu
859095
ileileargargalaalaaspleuilelysalavalprovalthrgln
100105110
asplysproleuglyleuglyhisalavalglyleualagluserval
115120125
leuaspaspaspgluaspvalvalalavalmetleuproaspaspleu
130135140
valleuprothrglyvalmetgluargmetalaglnvalargalaglu
145150155160
pheglyglyservalleucysalavalgluvalserglualaaspval
165170175
serlystyrglyilephegluileglualaaspthrlysaspserasp
180185190
vallyslysvallysglymetvalglulysproalailegluaspala
195200205
proserargleualaalathrglyargtyrleuleuasparglysile
210215220
pheaspalaleuargargilethrproglyalaglyglygluleugln
225230235240
leuthraspalaileaspleuleuileaspgluglyhisprovalhis
245250255
ilevalilehisglnglylysarghisaspleuglyasnproglygly
260265270
tyrileproalacysvalasppheglyleuserhisprovaltyrgly
275280285
alaglnleulysaspalailelysglnileleualagluhisgluala
290295300
alagluargilealaaspaspserglnvallys
305310315
<210>7
<211>315
<212>prt
<213>corynebacteriumglutamicumatcc13032
<400>7
metserleuproileaspgluhisvalasnalavallysthrvalval
151015
valproalaalaglyleuglythrargpheleuproalathrlysthr
202530
valprolysgluleuleuprovalvalaspthrproglyilegluleu
354045
ilealaalaglualaalagluleuglyalathrargleualaileile
505560
thralaproasnlysalaglyvalleualahisphegluargserser
65707580
gluleuglugluthrleumetgluargglylysthraspglnvalglu
859095
ileileargargalaalaaspleuilelysalavalprovalthrgln
100105110
asplysproleuglyleuglyhisalavalglyleualagluserval
115120125
leuaspaspaspgluaspvalvalalavalmetleuproaspaspleu
130135140
valleuprothrglyvalmetgluargmetalaglnvalargalaglu
145150155160
pheglyglyservalleucysalavalgluvalserglualaaspval
165170175
serlystyrglyilephegluileglualaaspthrlysaspserasp
180185190
vallyslysvallysglymetvalglulysproalailegluaspala
195200205
proserargleualaalathrglyargtyrleuleuasparglysile
210215220
pheaspalaleuargargilethrproglyalaglyglygluleugln
225230235240
leuthraspalaileaspleuleuileaspgluglyhisprovalhis
245250255
ilevalilehisglnglylysarghisaspleuglyasnproglygly
260265270
tyrileproalacysvalasppheglyleuserhisprovaltyrgly
275280285
alaglnleulysaspalailelysglnileleualagluhisgluala
290295300
alagluargilealaaspaspserglnvallys
305310315
<210>8
<211>387
<212>prt
<213>corynebacteriumglutamicumatcc13032
<400>8
metlysilealavalalaglyleuglytyrvalglyleuserasnala
151015
alaleuleuserlysasnhislysvalvalalavalaspileaspglu
202530
gluargvallysleuvalglnglupheargserproilevalaspser
354045
aspleugluglutyrleuserthrlysproglnasnleuthralathr
505560
thraspalaglualaalatyrlysglyalaasppheilevalileala
65707580
thrprothrasntyraspprogluserasnphepheaspthrserser
859095
valgluservalilegluilevalleulysvalserproglyserthr
100105110
ilevalilelysserthrileprovalglyphethrsergluleuarg
115120125
ilelyshisproglualaserileilepheserproglupheleuarg
130135140
gluglyargalaphetyraspasnleutyrproserargvalvalval
145150155160
glyaspargserproleuglyglugluphealathrleuleualaglu
165170175
glyalalysglulysproproileleuleuthraspserthrgluala
180185190
glualailelysleupheserasnthrtyrleualaleuargvalala
195200205
phepheasngluleuaspthrtyralaservalargserleuaspthr
210215220
lysglnileilegluglyvalglyleuaspproargileglyserhis
225230235240
tyrasnasnproserpheglytyrglyglytyrcysleuprolysasp
245250255
thrlysglnleuleualaasntyrlysaspvalproglnasnleuile
260265270
seralavalvalglnalaasnlysthrarglysasppheilealaglu
275280285
aspileleuserlysserprothrvalvalglyiletyrargleuval
290295300
metlysserglyseraspasnpheargserserserileglnglyval
305310315320
metlysargilelysalalysglyilegluilevalvalpheglupro
325330335
asnleuglyglugluthrphetyrasnserlysileleuasnaspile
340345350
glugluphelysasptyrcysaspileileilealaasnargprothr
355360365
aspgluleuseraspvalproglulysvaltyrthrargaspilephe
370375380
glnargasp
385
<210>9
<211>439
<212>prt
<213>corynebacteriumglutamicumatcc13032
<400>9
metargmetthrvalileglythrglytyrleuglyalathrhisala
151015
alacysmetalagluleuserhisgluvalleuglyvalaspvalasp
202530
glualalysilealaserleulysaspserlysvalprophepheglu
354045
proglyleuprogluvalleugluargasnleugluasnglyargleu
505560
asnphethrthrasptyralaglualaalaalaphealaglnvalhis
65707580
pheleuglyvalglythrproglnglnlysglythrtyralaalaasp
859095
leuthrtyrvalargglnvalvalgluaspleuvalproleuleuglu
100105110
glygluhisileilepheglylysserthrvalprovalglythrala
115120125
gluglnleuglngluleualaaspserleuvallysproglyserhis
130135140
valgluilealatrpasnproglupheleuarggluglytyralaval
145150155160
lysaspthrilethrproaspargilevalvalglyvalargglugly
165170175
alathralaglualailealaarggluvaltyralathralaileala
180185190
alaaspthrpropheleuvalthraspleualathralagluleuval
195200205
lysvalseralaasnalapheleualathrlysileserpheileasn
210215220
alavalalagluilecysgluglnthrglyalaaspvalvalalaleu
225230235240
alaaspalaileglyhisaspaspargileglyarglyspheleugly
245250255
alaglyleuglypheglyglyglycysleuprolysaspileargala
260265270
phemetalaargalaglygluleuglyalaaspglnalaleuthrphe
275280285
leuarggluvalaspserileasnmetargargargaspargvalval
290295300
glnleualalysglumetcysglyglyserleuleuglylysargval
305310315320
thrvalleuglyalaalaphelysproasnseraspaspvalargasp
325330335
serproalaleuservalalaglyserleuserleuglnglyalaala
340345350
valservaltyraspproglualametaspasnalaargargvalphe
355360365
prothrleusertyralaserserthrlysglualaleuileaspala
370375380
hisleuvalvalleualathrglutrpglnglupheargaspleuasp
385390395400
progluvalalaglyglyvalvalglulysargalaileileaspgly
405410415
argasnvalleuaspvalalalystrplysalaalaglytrpglumet
420425430
glualaleuglyargasnleu
435
<210>10
<211>29
<212>dna
<213>人工序列
<400>10
cgactgcacggtgcaccaatgcttctggc29
<210>11
<211>33
<212>dna
<213>人工序列
<400>11
catggtctgtttcctgtgtgaaattgttatccg33
<210>12
<211>47
<212>dna
<213>人工序列
<400>12
tttcacacaggaaacagaccatggttttcaagggtgaaaaatgaata47
<210>13
<211>29
<212>dna
<213>人工序列
<400>13
ttaaatataaccatttgggtttttcattt29
<210>14
<211>51
<212>dna
<213>人工序列
<400>14
aaaacccaaatggttatatttaaagattaggttgaaataatatgagtattc51
<210>15
<211>46
<212>dna
<213>人工序列
<400>15
agaggttccccggctttcggttttataaatcattctctattttttc46
<210>16
<211>46
<212>dna
<213>人工序列
<400>16
cgaattcgagctcggtacccggggtgctcatctagcatctggcttg46
<210>17
<211>24
<212>dna
<213>人工序列
<400>17
taaaaggggctatcattcggaccc24
<210>18
<211>22
<212>dna
<213>人工序列
<400>18
aaccgaaagccggggaacctct22
<210>19
<211>46
<212>dna
<213>人工序列
<400>19
aacgacggccagtgccaagcttagctcgtcctcattgatcagttcg46
<210>20
<211>25
<212>dna
<213>人工序列
<400>20
ggcactcgtagaacgcatcaatggt25
<210>21
<211>27
<212>dna
<213>人工序列
<400>21
caatcgttgcccaggagaaatcattga27
<210>22
<211>25
<212>dna
<213>人工序列
<400>22
ggataacaatttcacacaggaaaca25
<210>23
<211>25
<212>dna
<213>人工序列
<400>23
gtgaaaagaaaaaccaccctggcgc25
<210>24
<211>47
<212>dna
<213>人工序列
<400>24
gcgccagggtggtttttcttttcacctgagtcttagattttgagaaa47
<210>25
<211>21
<212>dna
<213>人工序列
<400>25
aacaagagaccgccaagggtg21
<210>26
<211>44
<212>dna
<213>人工序列
<400>26
cacccttggcggtctcttgttgtactcttggttccatgagtttg44
<210>27
<211>43
<212>dna
<213>人工序列
<400>27
gcaaactcatggaaccaagagtaccttagccttcctggttgtg43
<210>28
<211>25
<212>dna
<213>人工序列
<400>28
ggtactcttggttccatgagtttgc25
<210>29
<211>48
<212>dna
<213>人工序列
<400>29
tgtttcctgtgtgaaattgttatccctattttacttgagaatcgtctg48
<210>30
<211>54
<212>dna
<213>人工序列
<400>30
gaccatggaattcgagctcggtaccaagttgttgtaaagtcatgcgcatgtgtg54
<210>31
<211>26
<212>dna
<213>人工序列
<400>31
ttattcgacggtgacagactttgcca26
<210>32
<211>58
<212>dna
<213>人工序列
<400>32
tggcaaagtctgtcaccgtcgaataaggccgtgcataattaatcgcatgactcgacta58
<210>33
<211>47
<212>dna
<213>人工序列
<400>33
caagcttgcatgcctgcaggtcgacttagacttctgcaaccactgca47
<210>34
<211>11
<212>prt
<213>人工序列
<400>34
glyglyserglyglyserglyglyglyglyser
1510
<210>35
<211>33
<212>dna
<213>人工序列
<400>35
ggtggaagtggaggtagtggaggtggaggtagt33
<210>36
<211>33
<212>dna
<213>人工序列
<400>36
ggaggtagcggtggaagcggtggaggtggaagc33
- 还没有人留言评论。精彩留言会获得点赞!