一种高通量基因表达谱检测试剂盒的制作方法
本发明涉及一种高通量基因表达谱检测试剂盒及使用方法,适用于任何具有细胞基因表达水平检测需求的样本,涵盖任何真核生物的细胞。属于生物医学领域。
背景技术:
核酸是遗传信息的载体,是最重要的生物信息分子,是分子生物学研究的主要对象。随着精准医疗计划的进展,高通量基因测序技术将在以后的疾病精确诊断、疾病亚型分类、精准监控疾病发生发展、精准指导用药、特异疾病相关基因组数据库建立等方面发挥越来越关键作用。高通量测序的实现离不开样本的收集及测序前样本准备工作。尽管样本从收集到建库的成本显著降低,然而一旦涉及高通量、大量样本的建库,成本依然较高。目前商品化的转录组文库构建试剂盒几乎为进口试剂盒,价格昂贵,且试剂盒中各种成分相对独立,操作繁琐。因此,本发明提供了一种高通量基因表达谱检测试剂盒及使用方法,将整个样本建库前的所有处理步骤模块化,实现了样本处理的简易化、低起始量化、低成本化、高通量化。
技术实现要素:
本发明的试剂盒主要用于高通量的基因表达谱的检测,该检测试剂盒提供了从样本收集到mrna测序前的所有实验步骤所需的试剂及实验方法;一共可分为四个部分:1.逆转录反应模块,核酸纯化模块,双链cdna合成模块,测序文库构建模块。
本发明采用的技术方案为:
一种高通量基因表达谱检测试剂盒,该试剂盒包括逆转录反应模块、核酸纯化模块、双链cdna合成模块、测序文库构建模块;
该逆转录反应模块,包括细胞裂解液、逆转录mix、barcode板(即逆转录引物板)、备用96孔板;其中:
该逆转录反应模块,包括细胞裂解液、逆转录mix、barcode板、备用96孔板;其中:
细胞裂解液:用pbs配置的0.3%triton-100;
逆转录mix:0.5μldntp,0.25μl逆转录酶,1μl5×逆转录酶buffer[250mmtris-hcl(ph8.3),375mmkcl,15mmmgcl2],0.125μldtt,0.125μlrna酶抑制剂,1μlbetaine(5m),0.045μlmgcl2;
barcode板:含96种不同标签的逆转录引物(浓度为20μm):(5’—3’)
acactctttccctacacgacgctcttccgatctctcacatttttttttttttttttttttttttttttt(seqidno:1)acactctttccctacacgacgctcttccgatcttactgctttttttttttttttttttttttttttttt(seqidno:2)acactctttccctacacgacgctcttccgatctatcagctttttttttttttttttttttttttttttt(seqidno:3)acactctttccctacacgacgctcttccgatctcgactatttttttttttttttttttttttttttttt(seqidno:4)acactctttccctacacgacgctcttccgatctcgtcattttttttttttttttttttttttttttttt(seqidno:5)acactctttccctacacgacgctcttccgatctctcgattttttttttttttttttttttttttttttt(seqidno:6)acactctttccctacacgacgctcttccgatcttagcactttttttttttttttttttttttttttttt(seqidno:7)acactctttccctacacgacgctcttccgatcttctagctttttttttttttttttttttttttttttt(seqidno:8)acactctttccctacacgacgctcttccgatctcacgtatttttttttttttttttttttttttttttt(seqidno:9)acactctttccctacacgacgctcttccgatctatgctctttttttttttttttttttttttttttttt(seqidno:10)acactctttccctacacgacgctcttccgatctgctacatttttttttttttttttttttttttttttt(seqidno:11)acactctttccctacacgacgctcttccgatctgcatcttttttttttttttttttttttttttttttt(seqidno:12)acactctttccctacacgacgctcttccgatctacatgctttttttttttttttttttttttttttttt(seqidno:13)acactctttccctacacgacgctcttccgatctacgtcatttttttttttttttttttttttttttttt(seqidno:14)acactctttccctacacgacgctcttccgatctcatgcttttttttttttttttttttttttttttttt(seqidno:15)acactctttccctacacgacgctcttccgatctctagtctttttttttttttttttttttttttttttt(seqidno:16)acactctttccctacacgacgctcttccgatcttgcatctttttttttttttttttttttttttttttt(seqidno:17)acactctttccctacacgacgctcttccgatctcatctgtttttttttttttttttttttttttttttt(seqidno:18)acactctttccctacacgacgctcttccgatctcagatctttttttttttttttttttttttttttttt(seqidno:19)acactctttccctacacgacgctcttccgatctagcacttttttttttttttttttttttttttttttt(seqidno:20)acactctttccctacacgacgctcttccgatctactcgatttttttttttttttttttttttttttttt(seqidno:21)acactctttccctacacgacgctcttccgatcttcacgttttttttttttttttttttttttttttttt(seqidno:22)acactctttccctacacgacgctcttccgatctgtctactttttttttttttttttttttttttttttt(seqidno:23)acactctttccctacacgacgctcttccgatcttctcagtttttttttttttttttttttttttttttt(seqidno:24)acactctttccctacacgacgctcttccgatcttcagactttttttttttttttttttttttttttttt(seqidno:25)acactctttccctacacgacgctcttccgatctgactcatttttttttttttttttttttttttttttt(seqidno:26)acactctttccctacacgacgctcttccgatctcacagttttttttttttttttttttttttttttttt(seqidno:27)acactctttccctacacgacgctcttccgatctacactgtttttttttttttttttttttttttttttt(seqidno:28)acactctttccctacacgacgctcttccgatctgcagtatttttttttttttttttttttttttttttt(seqidno:29)acactctttccctacacgacgctcttccgatctgctgattttttttttttttttttttttttttttttt(seqidno:30)acactctttccctacacgacgctcttccgatcttagtcgtttttttttttttttttttttttttttttt(seqidno:31)acactctttccctacacgacgctcttccgatctatgacgtttttttttttttttttttttttttttttt(seqidno:32)acactctttccctacacgacgctcttccgatctcgatgttttttttttttttttttttttttttttttt(seqidno:33)acactctttccctacacgacgctcttccgatctagatcgtttttttttttttttttttttttttttttt(seqidno:34)acactctttccctacacgacgctcttccgatcttacgagtttttttttttttttttttttttttttttt(seqidno:35)acactctttccctacacgacgctcttccgatctcgtagatttttttttttttttttttttttttttttt(seqidno:36)acactctttccctacacgacgctcttccgatcttgtacgtttttttttttttttttttttttttttttt(seqidno:37)acactctttccctacacgacgctcttccgatctatcgtgtttttttttttttttttttttttttttttt(seqidno:38)acactctttccctacacgacgctcttccgatctgtgcattttttttttttttttttttttttttttttt(seqidno:39)acactctttccctacacgacgctcttccgatctgagctatttttttttttttttttttttttttttttt(seqidno:40)acactctttccctacacgacgctcttccgatctctcacaatttttttttttttttttttttttttttttt(seqidno:41)acactctttccctacacgacgctcttccgatcttactgcatttttttttttttttttttttttttttttt(seqidno:42)acactctttccctacacgacgctcttccgatctatcagcatttttttttttttttttttttttttttttt(seqidno:43)acactctttccctacacgacgctcttccgatctcgactaatttttttttttttttttttttttttttttt(seqidno:44)acactctttccctacacgacgctcttccgatctcgtcaattttttttttttttttttttttttttttttt(seqidno:45)acactctttccctacacgacgctcttccgatctctcgaattttttttttttttttttttttttttttttt(seqidno:46)acactctttccctacacgacgctcttccgatcttagcacatttttttttttttttttttttttttttttt(seqidno:47)acactctttccctacacgacgctcttccgatcttctagcatttttttttttttttttttttttttttttt(seqidno:48)acactctttccctacacgacgctcttccgatctcacgtaatttttttttttttttttttttttttttttt(seqidno:49)acactctttccctacacgacgctcttccgatctatgctcatttttttttttttttttttttttttttttt(seqidno:50)acactctttccctacacgacgctcttccgatctgctacaatttttttttttttttttttttttttttttt(seqidno:51)acactctttccctacacgacgctcttccgatctgcatctatttttttttttttttttttttttttttttt(seqidno:52)acactctttccctacacgacgctcttccgatctacatgcatttttttttttttttttttttttttttttt(seqidno:53)acactctttccctacacgacgctcttccgatctacgtcaatttttttttttttttttttttttttttttt(seqidno:54)acactctttccctacacgacgctcttccgatctcatgctatttttttttttttttttttttttttttttt(seqidno:55)acactctttccctacacgacgctcttccgatctctagtcatttttttttttttttttttttttttttttt(seqidno:56)acactctttccctacacgacgctcttccgatcttgcatcatttttttttttttttttttttttttttttt(seqidno:57)acactctttccctacacgacgctcttccgatctcatctgatttttttttttttttttttttttttttttt(seqidno:58)acactctttccctacacgacgctcttccgatctcagatcatttttttttttttttttttttttttttttt(seqidno:59)acactctttccctacacgacgctcttccgatctagcactatttttttttttttttttttttttttttttt(seqidno:60)acactctttccctacacgacgctcttccgatctactcgaatttttttttttttttttttttttttttttt(seqidno:61)acactctttccctacacgacgctcttccgatcttcacgtatttttttttttttttttttttttttttttt(seqidno:62)acactctttccctacacgacgctcttccgatctgtctacatttttttttttttttttttttttttttttt(seqidno:63)acactctttccctacacgacgctcttccgatcttctcagatttttttttttttttttttttttttttttt(seqidno:64)acactctttccctacacgacgctcttccgatcttcagacatttttttttttttttttttttttttttttt(seqidno:65)acactctttccctacacgacgctcttccgatctgactcaatttttttttttttttttttttttttttttt(seqidno:66)acactctttccctacacgacgctcttccgatctcacagtatttttttttttttttttttttttttttttt(seqidno:67)acactctttccctacacgacgctcttccgatctacactgatttttttttttttttttttttttttttttt(seqidno:68)acactctttccctacacgacgctcttccgatctgcagtaatttttttttttttttttttttttttttttt(seqidno:69)acactctttccctacacgacgctcttccgatctgctgatatttttttttttttttttttttttttttttt(seqidno:70)acactctttccctacacgacgctcttccgatcttagtcgatttttttttttttttttttttttttttttt(seqidno:71)acactctttccctacacgacgctcttccgatctatgacgatttttttttttttttttttttttttttttt(seqidno:72)acactctttccctacacgacgctcttccgatctcgatgtatttttttttttttttttttttttttttttt(seqidno:73)acactctttccctacacgacgctcttccgatctagatcgatttttttttttttttttttttttttttttt(seqidno:74)acactctttccctacacgacgctcttccgatcttacgagatttttttttttttttttttttttttttttt(seqidno:75)acactctttccctacacgacgctcttccgatctcgtagaatttttttttttttttttttttttttttttt(seqidno:76)acactctttccctacacgacgctcttccgatcttgtacgatttttttttttttttttttttttttttttt(seqidno:77)acactctttccctacacgacgctcttccgatctatcgtgatttttttttttttttttttttttttttttt(seqidno:78)acactctttccctacacgacgctcttccgatctgtgcatatttttttttttttttttttttttttttttt(seqidno:79)acactctttccctacacgacgctcttccgatctgagctaatttttttttttttttttttttttttttttt(seqidno:80)acactctttccctacacgacgctcttccgatcttcagacgtttttttttttttttttttttttttttttt(seqidno:81)acactctttccctacacgacgctcttccgatctgactcagtttttttttttttttttttttttttttttt(seqidno:82)acactctttccctacacgacgctcttccgatctcacagtgtttttttttttttttttttttttttttttt(seqidno:83)acactctttccctacacgacgctcttccgatctacactggtttttttttttttttttttttttttttttt(seqidno:84)acactctttccctacacgacgctcttccgatctgcagtagtttttttttttttttttttttttttttttt(seqidno:85)acactctttccctacacgacgctcttccgatctgctgatgtttttttttttttttttttttttttttttt(seqidno:86)acactctttccctacacgacgctcttccgatcttagtcggtttttttttttttttttttttttttttttt(seqidno:87)acactctttccctacacgacgctcttccgatctatgacggtttttttttttttttttttttttttttttt(seqidno:88)acactctttccctacacgacgctcttccgatctcgatgtgtttttttttttttttttttttttttttttt(seqidno:89)acactctttccctacacgacgctcttccgatctagatcggtttttttttttttttttttttttttttttt(seqidno:90)acactctttccctacacgacgctcttccgatcttacgaggtttttttttttttttttttttttttttttt(seqidno:91)acactctttccctacacgacgctcttccgatctcgtagagtttttttttttttttttttttttttttttt(seqidno:92)acactctttccctacacgacgctcttccgatcttgtacggtttttttttttttttttttttttttttttt(seqidno:93)acactctttccctacacgacgctcttccgatctatcgtggtttttttttttttttttttttttttttttt(seqidno:94)acactctttccctacacgacgctcttccgatctgtgcatgtttttttttttttttttttttttttttttt(seqidno:95)acactctttccctacacgacgctcttccgatctgagctagtttttttttttttttttttttttttttttt(seqidno:96)
该核酸纯化模块包括纯化用超顺磁性磁珠;
该双链cdna合成模块包括:双链合成buffer(10x)、双链合成enzymemix;
双链合成buffer(10x):200mmtris-hcl,120mm(nh4)2so4,50mmmgcl2,1.6mmb-nad,1.9mmdntpseach,ph7.4
双链合成enzymemix:6,000units/mldnapolymerasei(e.coli)5,000units/mlrnaseh,25,000units/mle.colidnaligase,10mmtris-hcl(ph7.5),50mmkcl,1mmdtt,0.1mmedta,50%glycerol;
该测序文库构建模块包括片段化酶mix、中和buffer、引物1,引物2、pcrmix;其中
片段化酶mix:nexteraxt(illumina)试剂盒中的tagmentdnabuffer和amplificationtagmentmix的混合物;
中和buffer:nexteraxt(illumina)试剂盒中的ntbuffer;
引物1:
序列为5’-aatgatacggcgaccaccgagatctacactctttccctacacg-3’(seqidno:97),浓度为5μm;
引物2,含12种不同标签的引物,浓度为5μm:
5’-caagcagaagacggcatacgagattcgccttagtctcgtgggctcgg-3’(seqidno:98)
5’-caagcagaagacggcatacgagatctagtacggtctcgtgggctcgg-3’(seqidno:99)
5’-caagcagaagacggcatacgagatttctgcctgtctcgtgggctcgg-3’(seqidno:100)
5’-caagcagaagacggcatacgagatgctcaggagtctcgtgggctcgg-3’(seqidno:101)
5’-caagcagaagacggcatacgagataggagtccgtctcgtgggctcgg-3’(seqidno:102)
5’-caagcagaagacggcatacgagatcatgcctagtctcgtgggctcgg-3’(seqidno:103)
5’-caagcagaagacggcatacgagatgtagagaggtctcgtgggctcgg-3’(seqidno:104)
5’-caagcagaagacggcatacgagatcctctctggtctcgtgggctcgg-3’(seqidno:105)
5’-caagcagaagacggcatacgagatagcgtagcgtctcgtgggctcgg-3’(seqidno:106)
5’-caagcagaagacggcatacgagatcagcctcggtctcgtgggctcgg-3’(seqidno:107)
5’-caagcagaagacggcatacgagattgcctcttgtctcgtgggctcgg-3’(seqidno:108)
5’-caagcagaagacggcatacgagattcctctacgtctcgtgggctcgg-3’(seqidno:109)
pcrmix:166.5mmkcl,33.3mmtris-hcl(ph8.3),6.66mmmgcl2,1mmdntp,0.5u高保真dnapolymerase。
本发明还提供了上述高通量基因表达谱检测试剂盒的使用方法,该方法包括如下步骤:
1)、逆转录反应
(1)、提前将96孔板低温离心机降温到4℃;
(2)当所收集的样本为体内组织或个体,则需用传统的trizol法提取总rna或直接通过任何商业化rna提取试剂盒提取rna,当所收集样本为体外细胞,则向样本中直接加入细胞裂解液对细胞进行裂解;
(3)每个样本取裂解后的样本2ul(rna总量为1-1000ng)于备用96孔板中,用排枪向每个孔加入0.5ul不同的逆转录引物(即barcode板中引物)。
(3)涡旋震荡混匀,3000rpm,4℃,离心3min,在pcr仪上反应(72℃,3min)后,立即放置在冰上;
(5)每个孔依次加入2.5μl逆转录mix
(6)涡旋震荡混匀,3000rpm,4℃,离心3min,在pcr仪上反应{42℃,90min;[50℃,2min;42℃,2min]10个循环,70℃,15min;16℃,5min;}
(7)逆转录后,同一组barcode的样本可混合到一个ep管中;
所有步骤均在冰上操作。
2)核酸纯化
(1)涡旋震荡纯化磁珠(提前将纯化磁珠平衡至室温),将磁珠混匀,并取90%体积的磁珠至逆转录混合产物中,涡旋震荡或使用移液器轻轻吹打至充分混匀;
(2)室温孵育5min;
(3)将ep管短暂离心并置于磁力架上约5min,用于分离磁珠和液体;待溶液澄清后,小心移除上清;
(4)保持ep管置于磁力架上,加入180ul-200ul新鲜配置70%~80%酒精漂洗磁珠,室温孵育30s,小心移除上清;
(5)重复(4)的漂洗步骤,总共漂洗2次;
(6)ep管开盖于空气中干燥5~10分钟至无酒精残留;
(7)加入适量灭菌双蒸水,涡旋振荡或用移液器轻轻吹打使与磁珠充分混匀后,室温孵育2min;
(8)将ep管置于磁力架上静置约5min,待溶液澄清后,小心移除上清至新的ep管中,移除过程中切勿触碰磁珠。
纯化后的样本4℃可储存一周,长期储存需放-20℃,避免反复冻融
3)、双链cdna合成
(1)取100ng上述混合、纯化后的cdna样本,依次加入2μl双链合成buffer,1μl双链合成enzymemix,并加水至20μl体系,涡旋震荡混匀;
(2)3000rpm,4℃,离心3min后,在pcr仪上反应(16℃,2.5h);
(3)按上述pcr反应产物按照核酸纯化模块步骤将双链合成产物进行纯化
所有步骤均在冰上操作;
4)、测序文库构建
(1)根据qubit检测的样本浓度,将其稀释至0.5-1ng/μl,取1.25μl,加入3.75μl片段化酶mix;
(2)涡旋20s,3000g离心5min后,在pcr仪上反应[55℃,10min;10℃,5min];
(3)等温度降到10℃后,迅速在反应体系中加入1.25μl中和buffer终止反应;涡旋20s,3000g离心5min;
(4)在上述反应体系中依次加入:3.75μlpcrmix,1.25μl的i5端富集引物,(引物1),1.25μli7端引物(引物2)
(5)涡旋20s,3000g离心2min后,在pcr仪上反应[72℃,3min;95℃,30s;12循环(95℃,10s;55℃,30s;72℃,60s);72℃,5min;10℃,5min;]
(6)按上述pcr反应产物按照核酸纯化模块步骤进行纯化;
(7)质检:将纯化后的洗脱产物,用qubit和agilentbioanalyzer2200检测样本中双链dna的浓度、碱基序列长度分布情况
所有步骤均在冰上操作,纯化后的样本4℃可储存一周,长期储存需放-20℃,避免反复冻融。
本发明所具有的有益效果:
本发明提供了一种高通量基因表达谱检测试剂盒及使用方法,将整个样本建库前的所有处理步骤模块化,实现了样本处理的简易化、低起始量化、高通量化、低成本化。本发明试剂盒能广泛应用于药物、生物材料等物理化学生物因素对体外细胞、体内组织、甚至个体的全基因表达谱的系统性检测。
附图说明
图1为逆转录反应模块流程图;
图2为核酸纯化模块流程图;
图3为双链cdna合成模块流程图;
图4为测序文库构建流程图;
图5为文库质检结果图;
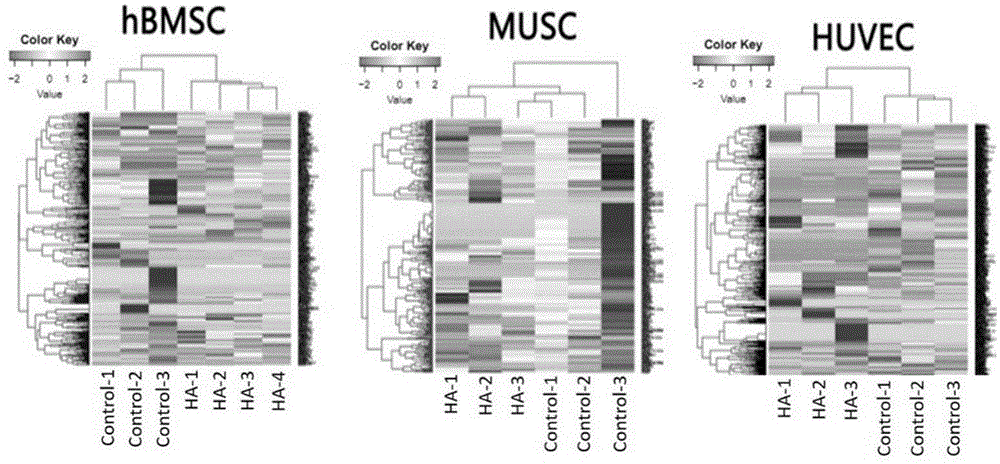
图6为体外细胞的高通量基因表达谱检测结果分析图;
图7为体内多组织的高通量基因表达谱检测结果分析图;
图8为单个线虫个体的高通量基因表达谱检测结果分析图。
具体实施方式
下面结合具体实施例对本发明进行进一步描述,但本发明的保护范围并不仅限于此:
实施例1:体外细胞的高通量基因表达谱检测
该实施例的实验目的为用ha纳米材料处理体外培养的hbmsc,myoblast,huvec三种细胞,用于检测和比较这该材料对这三种细胞的基因表达谱的影响。每种细胞分为两个组:材料处理组和非处理组,每个组设三个重复。总的细胞样本数为18个。
1)收集的细胞样本加入适量细胞裂解液;
2)每个样本取裂解后的样本2ul(rna总量为1-1000ng)于备用96孔板中,用排枪向每个孔加入0.5ul不同的逆转录引物(即barcode)。
3)涡旋震荡混匀,3000rpm,4℃,离心3min,在pcr仪上反应(72℃,3min)后,立即放置在冰上
4)每个孔依次加入2.5μl逆转录mix
5)涡旋震荡混匀,3000rpm,4℃,离心3min,在pcr仪上反应{42℃,90min;[50℃,2min;42℃,2min]10个循环,70℃,15min;16℃,5min;}
6)逆转录后,18个样本混合到一个ep管中。
7)纯化磁珠放置室温后,涡旋震荡纯化磁珠,将磁珠混匀,并取90%体积的磁珠至逆转录混合产物中,涡旋震荡或使用移液器轻轻吹打至充分混匀
8)室温孵育5min
9)将ep管短暂离心并置于磁力架上约5min,用于分离磁珠和液体;待溶液澄清后,小心移除上清
10)保持ep管置于磁力架上,加入180ul-200ul新鲜配置70%~80%酒精漂洗磁珠,室温孵育30s,小心移除上清
11)重复(10)的漂洗步骤,总共漂洗2次
12)ep管开盖,于空气中干燥5~10分钟至无酒精残留
13)加入适量灭菌双蒸水,涡旋振荡或用移液器轻轻吹打使与磁珠充分混匀后,室温孵育2min
14)将ep管置于磁力架上静置约5min,待溶液澄清后,小心移除上清至新的ep管中,移除过程中切勿触碰磁珠
15)纯化后的产物进行qubit测量浓度。
16)取100ng上述混合、纯化后的cdna样本,依次加入2μl双链合成buffer,1μl双链合成enzymemix,并加水至20μl体系,涡旋震荡混匀。
17)3000rpm,4℃,离心3min后,在pcr仪上反应(16℃,2.5h)。
18)将上述pcr反应产物按照核酸纯化模块步骤将双链合成产物进行纯化。
19)根据qubit检测的样本浓度,将其稀释至0.5-1ng/μl,取1.25μl,加入3.75μl片段化酶mix
20)涡旋20s,3000g离心5min后,在pcr仪上反应[55℃,10min;10℃,5min],等温度降到10度后,迅速在反应体系中加入1.25μl中和buffer终止反应。涡旋20s,3000g离心5min
21)在上述反应体系中依次加入:3.75μlpcrmix,1.25μl的i5端富集引物,(引物1),1.25μli7端引物(引物2)
22)涡旋20s,3000g离心2min后,在pcr仪上反应[72℃,3min;95℃,30s;12循环(95℃,10s;55℃,30s;72℃,60s);72℃,5min;10℃,5min;]
23)按上述pcr反应产物按照核酸纯化模块步骤进行纯化
24)质检:将纯化后的洗脱产物,用qubit和agilentbioanalyzer2200检测样本中双链dna的浓度、序列长度分布情况,结果显示符合测序上机要求(图5)
25)将高通量测序获得的数据利用r语言进行标准化、差异基因表达及聚类分析,发现同一处理组的重复样本的基因表达谱相似,并且两个比较组在基因表达上确实存在明显的差异(图6)。
实施例2:体内组织的高通量基因表达谱检测
该实施例的实验目的为用羟基磷灰石纳米材料对小鼠进行尾静脉注射,取小鼠心、肝、肾等7种组织和器官进行基因表达谱检测,用于检测和比较这该材料对小鼠体内组织的基因表达谱的影响。小鼠分为两个组:材料处理组和非处理组,每个组设三个重复。总的组织样本数为42个。
收集的42个组织样本用传统trizol法提取总rna,逆转录,核酸纯化,双链合成和文库构建步骤同上述18个体外细胞样本的方法。
该42个样本构建的文库进行高通量测序,获得的数据利用r语言进行标准化、差异基因表达及聚类分析,发现每一中组织与其它组织相比,组织基因表达谱相似,pca聚类分析可聚成一类,同时同一组织的两个比较组之间基因表达上确实存在明显的差异(如图7)
实施例3:个体的高通量基因表达谱检测
该实施例的实验目的为取不同温度处理(变频温度variable组,正常温度control组)的线虫进行线虫个体水平的基因表达谱检测,用于检测和比较不同温度对线虫基因表达谱的影响。分为两个组:变频温度组和正常温度组,每一条线虫作为一个样本,变频温度组19条,常温组有7条。即总的样本数为26个。
收集的26个组织样本用传统trizol法提取总rna,逆转录,核酸纯化,双链合成和文库构建步骤同上述18个体外细胞样本的方法。
将该26条线虫样本构建的文库进行高通量测序,获得的数据利用r语言进行标准化、差异基因表达及聚类分析,发现线虫样本的基因表达谱比较分散,pca聚类分析没有明显的聚类,但是两个比较组之间基因表达上确实存在明显的差异(如图8)。
- 还没有人留言评论。精彩留言会获得点赞!
- 一种构建Hi‑C高通量测序文库的方法与制造工艺
- 桑葚病原菌高通量鉴定及种属分类方法及其应用与制造工艺
- 基于高通量测序技术检测乳腺癌/卵巢癌易感基因的多重PCR特异性引物、试剂盒及方法与制造工艺
- 基于高通量测序检测人EGFR基因变异的质控方法及试剂盒与制造工艺
- 基于高通量测序技术的小麦病原微生物检测方法及其应用与制造工艺
- 一种基于二代高通量测序技术的地中海贫血症的基因检测试剂盒的制造方法与工艺
- 一种基于集群的高通量数据分析方法与制造工艺
- 高通量双折射干涉成像光谱仪装置及其成像方法与制造工艺
- 高通量测序检测人循环肿瘤DNA EGFR基因的捕获探针及试剂盒的制造方法与工艺
- 一种高通量测序检测无创亲子鉴定的方法与制造工艺