基于CRISPR的组合物和使用方法与流程
相关申请的交叉引用
本申请依据35u.s.c.119要求2014年12月18日和2015年10月9日提交并且名称为“基于crispr的组合物和使用方法(crispr-basedcompositionsandmethodsofuse)”的美国临时专利申请序列号62/093,588和62/239,546的优先权,所述临时专利申请的内容通过引用整体并入本文。
序列表
本申请包含序列表,其已经通过efs-web以ascii格式提交并且通过引用整体并入本文。ascii拷贝创建于2015年12月18日,命名为idt01-008-us_st25.txt,并且大小为177,163字节。
本发明涉及用于crispr系统中的修饰的组合物,和其使用方法。
背景技术:
使用成簇的规律间隔的短回文重复序列(crispr)和相关的cas蛋白(crispr-cas系统)进行位点特异性dna切割对于许多生物学应用显示出巨大的潜力。crispr被用于基因组编辑;转录阻遏因子(crispri)和激活因子(crispra)的基因组规模特异性靶向内源基因;以及利用cas酶的rna向导dna靶向的其它应用。
crispr-cas系统产生自细菌和古细菌,以提供针对病毒和质粒的适应性免疫。有三类可能潜在适用于研究和治疗性试剂的crispr-cas系统,但是ii型crispr系统在具有具有理想特征,利用单一crispr相关(cas)核酸酶(特别是cas9)与适当的向导rna的复合物(与包含crispr激活rna:反式激活crrna(crrna:tracrrna)对的细菌中的天然复合物类似的2部分rna系统或人工嵌合单向导rna(sgrna))介导目标dna的双链切割。在哺乳动物系统中,已通过在体外转录后转染含有驱动rna转录的rnapoliii启动子(例如u6或h1)、病毒载体和单链rna的dna盒,引入了这些rna(参见xu,t.等,《应用与环境微生物学(applenvironmicrobiol)》,2014.80(5):第1544-52页)。
在crispr-cas9系统中,使用例如以化脓性链球菌(streptococcuspyogenes)中存在的系统为例(s.py.或spy),天然crrna长度为约42bp,含有与目标序列(也称为前间区序列(protospacer))互补的约20个碱基的5'区和通常约22个碱基长度的对应于tracrrna序列的互补区的3'区。天然tracrrna长度为约85-90个碱基,具有含有与crrna互补的区域的5'区以及5'上游的约10个碱基区。tracrrna的其余3'区包括二级结构(在本文中称为“tracrrna3'尾”)。
jinek等广泛研究了crispr-cas9系统正常运作所需的crrna和tracrrna的部分(《科学(science)》,2012.337(6096):第816-21页)。他们设计了仍可在crispr-cas9中起作用的截短的crrna:tracrrna片段,其中crrna是野生型42个核苷酸并且tracrrna被截短到75个核苷酸。他们还开发了一种实施方案,其中crrna和tracrrna利用接头环连接,形成单向导rna(sgrna),其在不同的实施方案中在99-123个核苷酸之间变化。天然的2部分crrna:tracrrna复合物的构型示于图1中,并且人工sgrna单向导的99个核苷酸实施方案示于图2中。
至少两个团队已经阐明了化脓性链球菌(streptococcuspyogenes)cas9(spycas9)的晶体结构。在jinek,m.等中,该结构没有显示与向导rna或目标dna复合的核酸酶。他们进行了分子模拟实验,以揭示复合物形式的蛋白质与rna和dna之间的预测的相互作用(《科学(science)》,2014.343,第1215页,doi:10.1126/science/1247997)。
在nishimasu,h.等中,spycas9的晶体结构在2.5埃分辨率下显示为与sgrna和其目标dna的复合物(《细胞(cell)》,2014.156(5):第935-49页,整体并入本文)。晶体结构鉴定了cas9酶的两个叶:识别叶(rec)和核酸酶叶(nuc)。sgrna:目标dna异源双链体(带负电荷)位于两个叶之间的带正电荷的沟槽中。显示与已知蛋白质无结构相似性且因此可能是cas9特异性功能结构域的rec叶与彼此互补的crrna和tracrrna的部分相互作用。
另一个团队briner等(《分子细胞(molcell)》,2014.56(2):第333-9页,整体并入本文)鉴定并表征了天然crrna:tracrrna双链体和sgrna内的六个保守组件。
crispr-cas9系统用于基因组工程,如下:crrna的一部分与目标序列杂交,tracrrna的一部分与crrna的一部分杂交,并且cas9核酸酶与整个构建体结合并引导切割。cas9含有与核酸内切酶hnh和ruvc同源的两个结构域,其中hnh结构域切割与crrna互补的dna链,并且ruvc样结构域切割非互补链。这导致基因组dna中的双链断裂。当通过非同源末端连接(nhej)修复时,断裂通常偏移1个或更多个碱基,导致天然dna序列的破坏,并且在许多情况下,如果该事件发生在蛋白质编码基因的编码外显子中,则导致移码突变。断裂也可以通过同源依赖性重组(hdr)修复,这允许经由实验操作将新的遗传物质插入到由cas9切割产生的切割位点中。
用于将向导rna递送到哺乳动物细胞中的一些当前方法包括转染含有用于内源转录的rnapoliii启动子的双链dna(dsdna),病毒递送,转染作为体外转录(ivt)产物的rna,或显微注射ivt产物。这些方法中的每一种都有缺点。已知引入哺乳动物细胞中的未经修饰的外源rna通过toll样受体(tlr)、rig-i、oasi和识别病原体相关分子模式(pamp)的其它受体的识别启动先天免疫应答。然而,在大多数已公布的研究中,将通过t7rna聚合酶体外转录(ivt)的rna递送至细胞。这种类型的rna有效载荷(payload)已被证明是先天免疫应答的触发因子。上述替代性递送方法也各自具有其自身缺点。例如,dsdna盒可以导致整合,由rnapolii启动子内源驱动的向导rna转录可以组成性地持续存在,并且所转录的rna量是不可控制的。
rna由血清和细胞中存在的核酸酶快速降解。通过ivt方法或化学合成制备的未经修饰的crisprrna触发因子(crrna、tracrrna和sgrna)在递送至哺乳动物细胞期间或递送之后迅速降解。如果rna被化学修饰以获得核酸酶抗性,则将实现更大的活性。血清和细胞中存在的最有效的降解活性是3'-核酸外切酶(eder等,《反义研究和开发(antisenseresearchanddevelopment)》,1:141-151,1991)。因此,对合成寡核苷酸进行“封端”通常改进核酸酶稳定性。单链反义寡核苷酸(aso)和双链小干扰rna(sirna)的化学修饰已经得到充分研究,并且如今实践中有成功的方法(关于综述,参见:kurreck,《欧洲生物化学杂志(eur.j.biochem.)》,270:1628-1644,2003;behlke,《寡核苷酸(oligonucleotides)》,18:305-320,2008;lennox等,《基因疗法(genetherapy)》,18:1111-1120,2011)。因此,需要制定供crispr/cas的rna组分使用的化学修饰策略。虽然可用的化学修饰的基本工具箱是本领域技术人员熟知的,但是位点特异性修饰对rna物质和效应蛋白的相互作用的影响不容易预测,并且有效的修饰模式通常必须凭经验确定。在一些情况下,rna的序列可能影响修饰模式的有效性,需要调整用于不同序列背景的修饰模式,使得这些方法的实际应用更具挑战性。
因此,需要修饰向导rna以降低其对细胞的毒性并延长在哺乳动物细胞中的寿命和功能性,同时仍然在crispr-cas系统中执行其预期目的。本文所述的本发明的方法和组合物提供用于crispr-cas系统中的rna和修饰的rna寡核苷酸。根据本文提供的本发明的描述,本发明的这些和其它优点以及另外的发明特征将是显而易见的。
技术实现要素:
本发明涉及用于crispr系统中的修饰的组合物和其使用方法。所述组合物包括修饰的核苷酸间的键和2'-o-烷基和2'-o-氟修饰的rna寡核苷酸,以用作crispr-cas系统的向导链(crrna:tracrrna或sgrna)。组合物还包括末端修饰,例如阻止核酸外切酶攻击的反向dt(inverted-dt)碱基或其它非核苷酸修饰剂(例如丙二醇基团(c3间隔基)、萘基-偶氮修饰剂或如本领域中众所周知的其它修饰剂)。
在第一方面,提供了分离的tracrrna,其包括seqidno.:18的长度修饰形式。所述分离的tracrrna在成簇的规律间隔的短回文重复序列(crispr)-crispr相关(cas)(crispr-cas)核酸内切酶系统中显示活性。
在第二方面,提供了分离的crrna,其包括式(i)的长度修饰形式:
5'-x—z-3'(i),
其中x表示包含的靶特异性前间区序列结构域的序列,并且z表示包含通用的tracrrna结合结构域的序列,所述前间区序列结构域包含约20个靶特异性核苷酸,所述tracrrna结合结构域包含约20个核苷酸。所述分离的crrna在成簇的规律间隔的短回文重复序列(crispr)-crispr相关(cas)(crispr-cas)核酸内切酶系统中显示活性。
在第三方面,提供了分离的tracrrna,其包括seqidno.:2、18、30-33和36-39之一的化学修饰形式。所述分离的tracrrna在成簇的规律间隔的短回文重复序列(crispr)-crispr相关(cas)(crispr-cas)核酸内切酶系统中显示活性。
在第四方面,提供了分离的crrna,其包括式(i)的化学修饰形式:
5'-x—z-3'(i),
其中x表示包含靶特异性前间区序列结构域的序列,并且z表示包含通用的tracrrna结合结构域的序列,所述前间区序列结构域包含约17个核苷酸至约20个核苷酸,所述tracrrna结合结构域包含约12个核苷酸至约19个核苷酸。所述分离的crrna在成簇的规律间隔的短回文重复序列(crispr)-crispr相关(cas)(crispr-cas)核酸内切酶系统中显示活性。
附图说明
图1图示了具有42个碱基的未修饰的crrna(seqidno.:46)和89个碱基的未修饰的tracrrna(seqidno.:18)的野生型(wt)天然2部分crrna:tracrrna复合物。小写字母代表rna。
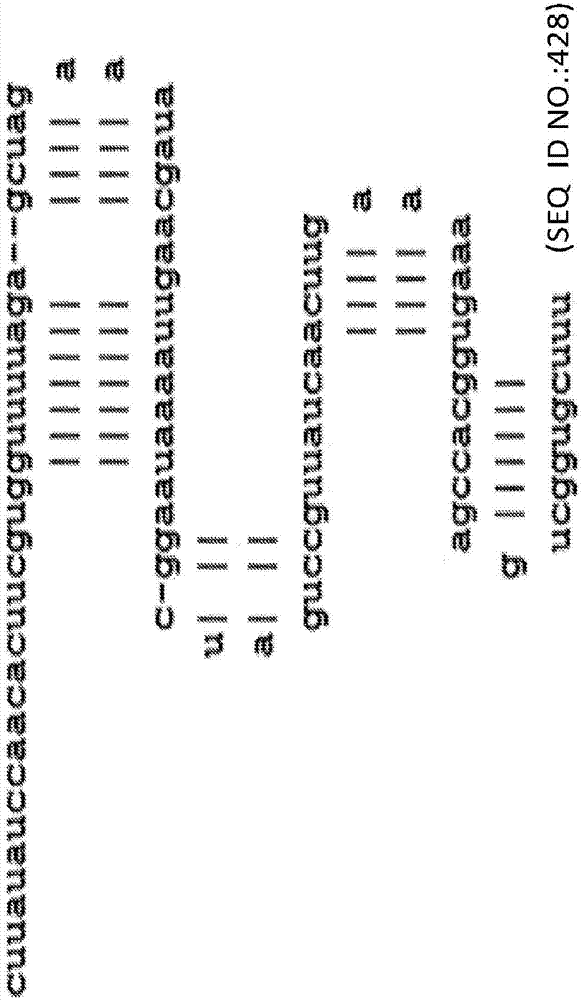
图2图示了通过添加新的发夹环将crrna和tracrrna元件融合成单个序列的99个碱基人工单向导rna(seqidno:428)(sgrna)。小写字母代表rna。
图3示出了实施例2中所研究的全长和截短的tracrrna种类的比对。序列是rna并且以5'-3'显示。比对基于上面的89个碱基wttracrrna序列(seqidno.:18)。内部空位代表内部截短/缺失的位点。大写字母代表rna。
图4示出了实施例3中所研究的全长和截短的crrna和tracrrna物质的比对。比对基于在其各自分组上面的42个碱基wtcrrna(seqidno.:46)和89个碱基wttracrrna(seqidno.:18)序列。crrna中的20个碱基的5'结构域具有序列特异性并且靶向人hprt1。3'结构域加有下划线并结合到tracrrna的5’末端的区域。tracrrna中的5'结构域加有下划线,其结合crrna的3’末端。大写字母代表rna。
图5图示了具有36个碱基crrna(seqidno.:48)和67个碱基tracrrna(seqidno.:2)的截短的2部分crrna:tracrrna复合物。小写字母代表rna。
图6的示意图示出了优化的截短和化学修饰的tracrrna(seqidno.:134)的一种实施方案的结构。长度为67个碱基。rna为小写字母并且2'omerna为大写字母。硫代磷酸酯(ps)核苷酸间的键由“*”指示。当从rna转化为2'omerna时导致功能显著损失的残基通过大箭头识别,并且当从rna转化为2'omerna时导致功能中度损失的残基通过小箭头识别。
图7的示意图示出了优化的截短和化学修饰的crrna(seqidno.:239)的一种实施方案的结构。长度为36个碱基。rna为小写字母并且2'omerna为大写字母。硫代磷酸酯(ps)核苷酸间的键由“*”指示。当从rna转化为2'omerna时导致功能显著损失的残基通过大箭头识别,并且当从rna转化为2'omerna时导致功能中度损失的残基通过小箭头识别。指出了5’末端20个碱基的前间区序列靶特异性向导结构域,其在这种情况下是对人hprt1基因具特异性的序列。指出了3’末端16个碱基的tracrrna结合结构域。
图8的示意图示出了实施例8中使用的优化的截短/修饰的crrna:tracrrna复合物的一种实施方案的结构。crrna位于顶部,其中5'-前间区序列结构域20个碱基加有下划线,其在这种情况下对目标人hprt1位点38285具特异性;3’末端是16个碱基的tracrrna结合结构域。下面比对tracrrna。rna为小写字母,2'omerna为大写字母,并且“*”指示硫代磷酸酯核苷酸间的键修饰。该图示出了由crrnaseqidno.:178和tracrrnaseqidno.:100形成的复合物。
图9的示意图示出了高度修饰的优化的截短/修饰的crrna:tracrrna复合物的一种实施方案的结构。crrna位于上面,其中5'-前间区序列结构域20个碱基加有下划线,其在这种情况下对目标人hprt1位点38285具特异性;3’末端是16个碱基的tracrrna结合结构域。下面比对tracrrna。rna为小写字母,2'omerna为大写字母,并且“*”指示硫代磷酸酯核苷酸间的键修饰。该图示出了由crrnaseqidno.:446和tracrrnaseqidno.:134形成的复合物。
图10的示意图示出了实施例10中使用的crrna修饰模式。寡核苷酸序列(分别是seqidno.:429-439,按出现次序)以5'-3'显示。小写字母=rna;加下划线=2'-o-甲基rna;c3=c3间隔基(丙二醇修饰剂);*=硫代磷酸酯核苷酸间的键;zen=萘基-偶氮修饰剂。指出了5'-靶特异性前间区序列结构域。碱基在该结构域中由“n”指示,因为每个目标位点的序列是不同的,但使用的修饰模式保持恒定。指出了3'通用tracrrna结合结构域。对修饰模式进行编号以供在表10与图10之间参考。
图11是表10中的数据的图(plot),其示出使用在人hprt1基因的12个不同位点测试的利用11种不同修饰模式制备的crrna在哺乳动物细胞中使用t7e1测定观察到的功能基因编辑。所有crrna变体都与优化的经修饰的tracrrna(seqidno.:100)配对。
图12的示意图示出了使用通用并可以应用于任何序列背景中的crrna修饰模式6进行高度修饰的优化的截短/修饰的crrna:tracrrna复合物的一种实施方案的结构。crrna(seqidno.:440)位于上面,其中5'-前间区序列结构域20个碱基加有下划线(n-碱基);3’末端是16个碱基的tracrrna结合结构域。下面比对tracrrna(seqidno.:134)。rna为小写字母,2'omerna为大写字母,并且“*”指示硫代磷酸酯核苷酸间的键修饰。
图13示出了来自用不同crisprgrna转染的hek-cas9细胞的rt-qpcr数据的图,其示出ifit1和ifitm1(涉及干扰素信号通路的2种基因)的相对表达水平。
发明详述
本发明的方面涉及用于crispr系统中的修饰的组合物和其使用方法。
如本文所用的术语“寡核苷酸”是指多脱氧核糖核苷酸(含有2-脱氧-d-核糖)、多核糖核苷酸(含有d-核糖)和并且任何其它类型的多核苷酸,其是嘌呤或嘧啶碱基的n-糖苷(单一核苷酸也称为“碱基”或“残基”)。术语“核酸”、“寡核苷酸”和“多核苷酸”之间在长度上没有任何预期区别,并且这些术语可以互换使用。这些术语仅指分子的主要结构。因此,这些术语包括双链和单链dna,以及双链和单链rna。为了用于本发明,寡核苷酸还可以包含其中碱基、糖或磷酸酯骨架被修饰的核苷酸类似物,以及非嘌呤或非嘧啶核苷酸类似物。寡核苷酸可包含核糖核苷酸、脱氧核糖核苷酸、修饰的核苷酸(例如具有2'修饰的核苷酸、合成碱基类似物等)或其组合。
本发明的组合物包括可能降低先天免疫系统激活的任何修饰。修饰可以在常规的磷酸二酯键处、核糖处或rna的核碱基处置换或取代。这样的组合物可以包括例如修饰的核苷酸,例如2'-o-甲基修饰的rna。
更广义地,术语“修饰的核苷酸”是指具有一个或多个对核苷、核碱基、戊糖环或磷酸基的修饰的核苷酸。例如,修饰的核苷酸不包括含有单磷酸腺苷、单磷酸鸟苷、单磷酸尿苷和单磷酸胞苷的核糖核苷酸,以及含有单磷酸脱氧腺苷、单磷酸脱氧鸟苷、单磷酸脱氧胸苷和单磷酸脱氧胞苷的脱氧核糖核苷酸。修饰包括由修饰核苷酸的酶(例如甲基转移酶)的修饰产生的天然存在的修饰。修饰的核苷酸还包括合成或非天然存在的核苷酸。修饰还包括碱基类似物和通用碱基。核苷酸中的合成或非天然存在的修饰包括具有2'修饰的那些,例如2'-o-烷基(包括2'-o-甲基)、2'-氟、2'-甲氧基乙氧基、2′-烯丙基、2′-o-[2-(甲基氨基)-2-氧代乙基]、4′-硫代、双环核酸,4′-ch2—o-2′-桥、4′-(ch2)2—o-2′-桥、2'-lna和2'-o-(n-甲基氨基甲酸酯)或包含碱基类似物的那些。这样的修饰基团描述于例如eckstein等,美国专利no.:5,672,695和matulic-adamic等,美国专利no.:6,248,878中。
已经在sirna文献(参见behlke,m.a.,《寡核苷酸(oligonucleotides)》,2008.18(4):第305-19页)以及mrna递送(参见sahin,u.等,《自然评论:药物发现(natrevdrugdiscov)》,2014.13(10):第759-80页)中记录了2'-o-甲基的使用。sahin等描述了延伸超出2'-ome修饰的mrna治疗剂的修饰和“非免疫原性”mrna。
术语“核糖核苷酸”涵盖天然的和合成的、未修饰的和修饰的核糖核苷酸。修饰包括糖部分的改变,碱基部分的改变和/或寡核苷酸中的核糖核苷酸之间的键的改变。
术语“cas9蛋白”涵盖当与合适的向导rna(例如sgrna或二元crrna:tracrrna组合物)组合形成活性crispr-cas核酸内切酶系统时具有生物化学和生物活性的野生型和突变形式的cas9。这包括在本发明中作为实例使用的具有来自化脓性链球菌cas9的不同氨基酸序列的直系同源物和cas9变体。
作为修饰rna的术语,术语“长度修饰”是指缺失核苷酸序列的参考rna的缩短或截短形式,或者包括额外的核苷酸序列的参考rna的延长形式。
作为术语修饰rna的术语“化学修饰”是指含有与rna共价连接的化学修饰核苷酸或非核苷酸化学基团的参考rna的形式。如本文所述的化学修饰的rna通常是指使用寡核苷酸合成程序制备的合成rna,其中修饰的核苷酸是在合成rna寡核苷酸期间引入的。然而,化学修饰的rna还包括在合成后用合适的修饰剂修饰的合成rna寡核苷酸。
申请人发现了在成簇的规律间隔的短回文重复序列(crispr)-crispr相关(cas)(crispr-cas)核酸内切酶系统中显示稳定活性的新颖crrna和tracrrna寡核苷酸组合物。所述寡核苷酸组合物包括crrna和tracrrna的长度修饰形式,以及crrna和tracrrna的化学修饰形式。crrna和tracrrna的长度修饰形式使得能够利用常规可用的具成本效益的和有效的寡核苷酸合成方案来制备这些rna的活性形式。crrna和tracrrna的化学修饰形式提供了具有某些特定性质可调节(如在细胞和体内背景中改进的稳定性)的活性剂。crrna和tracrrna的长度修饰形式也可以包括修饰,从而可以获得在crispr-cas核酸内切酶系统背景中具有活性的广泛多种组合物。下文描述这些寡核苷酸组合物和其在crispr-cas核酸内切酶系统中的性质。
crrna和tracrrna的长度修饰形式
图1描绘了野生型化脓链球菌(s.pyogenes)crrna:tracrrna复合物的图示,其中示例性的分离的crrna(seqidno.:46)与分离的tracrrna(seqidno.:8)配对。在第一方面,提供了包括seqidno.:18的长度修饰形式的分离的tracrrna。所述分离的tracrrna在crispr-cas核酸内切酶系统中显示活性。在一个方面,分离的tracrrna包括具有缺失序列信息的seqidno.:18核苷酸的长度修饰形式。在一些实施方案中,seqidno.:18的长度修饰形式包括seqidno.:18的缩短或截短形式,其中seqidno.:18可以在5’末端缩短1至20个核苷酸和在3’末端缩短1-10个核苷酸。当与crispr-cas核酸内切酶系统中功能合格的(competent)crrna配对时,seqidno.:18的这种缩短或截短形式保留活性。如果进行tracrrna的5’末端缩短并延伸到与crrna的3’末端配对的序列中,则可使用增强这些结构域中的结合亲和力的化学修饰来获得改进的活性。如果进行crrna的3’末端的缩短并延伸到与tracrrna的5’末端配对的序列中,则可使用增强这些结构域中的结合亲和力的化学修饰来获得改进的活性。具有缩短或截短形式的seqidno.:18的长度修饰形式的优选实例包括seqidno:2、30-33和36-39。具有缩短或截短形式的seqidno.:18的长度修饰形式的高度优选的实例包括seqidno:2。对于具有缩短或截短形式的seqidno.:18的上述示例性长度修饰形式中的每一个,seqidno.:2、30-33和36-69可以由化学上未修饰的核苷酸组成。
在第二方面,提供了包含式(i)的长度修饰形式的分离的crrna:
5'-x—z-3'(i),
其中x表示包括靶特异性前间区序列结构域的序列,并且z表示包括tracrrna结合结构域的序列。
靶特异性前间区序列结构域(式(i)的x结构域)通常包括与crispr-cas核酸内切酶系统靶向的dna区域互补的约20个核苷酸。在大多数crispr核酸内切酶系统中,tracrrna结合结构域(式(i)的z结构域)通常包括约20个核苷酸(在天然s.py.形式中,该结构域为22个核苷酸)。分离的crrna在crispr-cas核酸内切酶系统中显示活性。
在一个方面,分离的crrna包括具有缺失序列信息的式(i)的长度修饰形式。在一些实施方案中,式(i)的长度修饰形式包括式(i)的缩短或截短形式,其中式(i)可以在z结构域的3’末端缩短1-8个核苷酸。式(i)的长度修饰形式可以在x结构域的5端缩短以容纳具有17、18、19或20个核苷酸的靶特异性前间区序列结构域。式(i)的这种长度修饰形式的非常优选的实例包括具有19或20个核苷酸的靶特异性前间区序列结构域。具有长度为17-20个核苷酸的靶特异性前间区序列(x结构域)和/或在z结构域的3’末端缺失1-8个核苷酸的具有缩短或截短形式的式(i)的示例性长度修饰形式可以由化学上未修饰的核苷酸组成。
当与crispr-cas核酸内切酶系统中的合格的tracrrna配对时,式(i)的这种缩短或截短形式保留活性。具有式(i)的长度修饰形式的分离的式(i)的crrna的优选实施方案可以包括化学上未修饰的核苷酸和化学修饰核苷酸。
crrna和tracrrna的化学修饰形式
在第三方面,提供了包括化学修饰核苷酸或非核苷酸化学修饰剂的分离的tracrrna。分离的tracrrna在crispr-cas核酸内切酶系统中显示活性。在一个方面,分离的tracrrna包括具有选自核糖修饰、末端修饰基团和核苷酸间修饰的键的修饰的化学修饰核苷酸。示例性核糖修饰包括2'o-烷基(例如2'ome)、2'f、双环核酸和锁核酸(lna)。示例性末端修饰基团包括丙二醇(c3)间隔基和萘基-偶氮修饰剂(n,n-二乙基-4-(4-硝基萘-1-基偶氮)-苯胺或“zen”)和反向dt残基。示例性核苷酸间修饰的键包括硫代磷酸酯修饰。在一个方面,具有化学修饰形式的分离的tracrrna包括seqidno.:46和其长度修饰形式,例如seqidno.:46的缩短或截短形式。具有化学修饰核苷酸的seqidno.:46的优选的缩短或截短形式包括具有化学修饰核苷酸的seqidno:2、30-33和36-39。在实施例中呈现了在crispr-cas核酸内切酶系统中具有稳定活性的具有化学修饰核苷酸的分离的tracrrna的其它实例。
在第四方面,提供了包括化学修饰核苷酸的分离的crrna。所述分离的crrna在crispr-cas核酸内切酶系统中显示活性。在一个方面,分离的crrna包括具有选自核糖修饰、末端修饰基团和核苷酸间修饰的键的修饰的化学修饰核苷酸。示例性核糖修饰包括2'o-烷基(例如2'ome)、2'f、双环核酸和锁核酸(lna)。示例性末端修饰基团包括丙二醇(c3)间隔基和萘基-偶氮修饰剂(n,n-二乙基-4-(4-硝基萘-1-基偶氮)-苯胺或“zen”)和反向dt残基。示例性核苷酸间修饰的键包括硫代磷酸酯修饰。在一个方面,具有化学修饰形式的分离的crrna包括式(i)的crrna和其长度修饰形式。具有化学修饰核苷酸的式(i)的crrna的优选缩短或截短形式包括seqidno.:429-439。具有化学修饰核苷酸的分离的crrna的非常优选的实例包括seqidno.:434和435。这些特定的分离的crrna物质代表具有化学修饰核苷酸的“通用”crrna,当与crispr-cas核酸内切酶系统中的合格的tracrrna组合时显示高活性。在实施例中呈现了在crispr-cas核酸内切酶系统中具有稳定活性的具有化学修饰核苷酸的分离的crrna的其它实例。
上述分离的、长度修饰和化学修饰的crrna和tracrrna优选包括在2'-oh基团的化学修饰(例如,2'ome、2'f、双环核酸、锁核酸等)和封端修饰(例如,zen、c3间隔基、反向dt)。使用这两种类型的一般修饰给分离的、长度修饰和化学修饰的crrna和tracrrna提供了分离的、长度修饰和化学修饰的crrna和tracrrna在生物学背景下的生物化学稳定性和免疫耐受性。
上述分离的、长度修饰和化学修饰的crrna和tracrrna可以不同的组合混合,形成作为cas9的向导rna的活性crrna:tracrrna。例如,分离的长度修饰的tracrrna可以与分离的化学修饰的crrna组合以,形成作为cas9的向导rna的活性crrna:tracrrna。实施例提供了分离的、长度修饰和化学修饰的crrna和tracrrna的不同组合产生作为cas9的向导rna的活性crrna:tracrrna的说明。
需要在分离的、长度修饰和化学修饰的crrna和tracrrna中的一者(或两者)中包括特定的化学修饰核苷酸的程度取决于所得到的活性crrna:tracrrna用作cas9的向导rna的应用。在crispr-cas核酸内切酶系统的某些生物化学测定中,特别是在可以最小化或不存在核酸酶的情况下,可能不需要广泛地化学修饰的crrna和tracrrna来实现所得到的crispr-cas核酸内切酶系统的cas9的向导rna的稳定活性。这归因于如下事实:当可以使核酸酶最少或核酸酶不存在时,赋予核酸酶抗性的化学修饰核苷酸是不必要的。在某些生物学(体内)背景中,其中包括crrna和tracrrna的混合物在载体(如脂质体纳米颗粒)内被递送到细胞,与作为分离的“裸”rna混合物直接递送到血流中或注入器官系统中的crrna和tracrrna的混合物相比,分离的长度修饰和化学修饰的crrna和tracrrna可能需要较少的化学修饰的核苷酸。化学修饰的crrna和tracrrna中存在的化学修饰程度可以决定相关rna分子在体内(即在相关生物学背景下,例如在血流中或细胞内)的半衰期。因此,化学修饰的crrna和tracrrna的修饰概况可用于微调作为crispr-cas核酸内切酶系统中的cas9的向导rna的所得crrna:tracrrna双链体的生物化学和生物活性。
尽管现有技术关注于cas9与sgrna相互作用时的结构,但是所公开的本文所述的设计模式涵盖了上述crrna:tracrrna二元rna系统。单链向导rna提供几个益处,例如治疗设计的简单性。然而,标准固相亚磷酰胺rna合成显示,随着长度增加,寡核苷酸的产率降低,并且当长度超过60-70个碱基时,这个问题变得更加明显。这阻碍了一些tracrrna以及嵌合sgrna的稳定的具成本效益的合成,特别是在一些商业或治疗应用所需的较大规模下。因此,本发明不仅涵盖sgrna作为cas9的向导rna的实施方案,还涵盖替代的二元crrna:tracrrna作为cas9的向导rna的实施方案。然而,当与crispr-cas核酸内切酶系统中的cas9组合时,可以通过将适当的crrna和tracrrna连接或合成为基于本文提供的crrna和tracrrna的分离的、长度修饰和化学修饰形式的人工单分子sgrna来工程化具有稳定活性的分离的向导rna。可以通过直接合成或通过较短链的合成后化学结合来获得这种类型的长的单个向导物。
长度修饰和化学修饰的tracrrna组合物的设计解决了与长度>80个核苷酸的tracrrna寡核苷酸相关的潜在合成问题。2'-ome修饰的rna单体(在2'-oh上有效含有保护基团)的偶联效率大于rna单体偶联。并入2'-ome修饰的rna提供了一些优点。首先,它允许更长的寡核苷酸被合成为完整的2'-ome或rna/2'-ome混合寡核苷酸。其次,本发明的方法和组合物导致可以逃避免疫系统检测的crrna:tracrrna的合成和转染。众所周知,外源性未修饰的rna在哺乳动物细胞以及整个动物中触发先天免疫应答。使用2'ome修饰的寡核苷酸可以赋予rna对核酸酶的稳定性(第三个优点)以及减少与免疫原性触发因子相关的细胞死亡和毒性。这些优点不是2'-ome修饰本身所独有的,因为其它所公开的具有不同化学部分(例如2'f、其它2'o-烷基、lna和其它双环核苷酸)的修饰核苷酸可以在赋予核酸酶抗性方面提供类似的益处和优点。
在另一种实施方案中,与crrna互补的tracrrna部分含有至少一个修饰的核苷酸,并且在进一步的实施方案中,与crrna互补的tracrrna部分包含超过10%的修饰的残基,并且在进一步的实施方案中,与crrna不互补的tracrrna部分包含超过50%的修饰的残基,并且在进一步实施方案中与crrna不互补的tracrrna部分包含超过90%的修饰残基。
在另一种实施方案中,crrna部分是未修饰的,并且tracrrna部分包含至少一个修饰的核苷酸。在另一种实施方案中,crrna部分是未修饰的,并且tracrrna部分包含超过10%的修饰碱基。
在另一种实施方案中,分离的式(i)的crrna被设计为具有凭经验确定的修饰。如图7和图10中所描绘,在z结构域(tracrrna结合结构域)的3’末端的12个核苷酸和在x结构域的5’末端的10-12个核苷酸(在前间区序列结构域内)代表适于用化学修饰核苷酸取代的通用核苷酸,其中所得rna在crispr-cas核酸内切酶系统中保持稳定活性。在z结构域(tracrrna结合结构域)的5’末端内的其它核苷酸不耐受用化学修饰核苷酸取代(图7)。然而,分离的式(i)的crrna内的其它位点接受化学修饰核苷酸并在crispr-cas核酸内切酶系统中保留活性的能力在很大程度上凭经验确定。crrna的tracrrna结合结构域(z结构域)是恒定的(即,序列不会随着目标位点变化而改变),因此不管目标位点如何,本文描述的修饰模式对于所有crrna是通用的,并且可广泛应用。crrna的前间区序列(x结构域)随着靶而变化,并且该结构域内的一些碱基位置对化学修饰的耐受性随着序列背景而变化,并且如果需要位点的最大化学修饰,则可受益于经验优化。然而,靶特异性前间区序列(x)结构域内的一些残基可以在不考虑序列背景的情况下被修饰。该结构域的5’末端的10-12个残基可以被2'-修饰的残基取代,期望将维持修饰crrna的完全活性。根据序列背景,前间区序列(x)结构域的3’末端的其余8-10个碱基可能耐受修饰或可能不耐受修饰。前间区序列(x)结构域的20个碱基中的17个碱基可以在维持完全活性的同时进行修饰的一种序列背景示于图7中。指出了修饰损害活性的位点。
基于cas9的工具的应用是多种多样的。它们包括(但不限于):植物基因编辑、酵母基因编辑、敲除/敲入动物系的快速生成、生成疾病状态的动物模型、矫正疾病状态、插入报告基因以及全基因组功能性筛选。
本发明的实用性通过以下进一步扩展:包括cas酶的突变形式,例如cas9的d10a和h840a双突变体,作为与转录激活因子(crispra)和阻遏因子(crispri)的融合蛋白(参见xu,t.等,《应用与环境微生物学(applenvironmicrobiol)》,2014.80(5):第1544-52页)。cas9-sgrna复合物也可用于靶向单链mrna(参见o'connell,m.r.等,《自然(nature)》,516:263,2014)。以与靶向dsdna相同的方式,crrna:tracrrna可以与pammerdna寡核苷酸一起使用以将cas9切割引导至靶mrna,或者将其用于o'connell描述的mrna捕获测定中。
通过利用对于crispr/cas9应用递送合成rna寡核苷酸的方法,可以1)使用质谱法来确定离散的rna序列,2)将2'-ome修饰的rna选择性插入耐受良好的位置以赋予稳定性并避免免疫原性,但仍保留功能性效力,3)特异性控制引入细胞中的rna量以实现受控的瞬时效应,和4)消除关于引入的dsdna会被内源转录为rna但也可以成为同源性定向修复途径中或非同源末端连接中的底物从而导致整合事件的的担忧。这些整合事件可能导致crrna或tracrrna元件的长期的不期望的表达。此外,整合可能以随机和不可预测的方式破坏其它基因,从而以不期望的和潜在有害的方式改变细胞的遗传物质。因此,本发明希望作为在细胞中引入crispr途径的元件的瞬时表达的手段,引入的方式是瞬时的,并且不留下持续迹象或者改变在预期由crrna向导所引导的任何变异之外的基因组。
crispr-cas核酸内切酶系统
合格的crispr-cas核酸内切酶系统包括由分离的cas9蛋白和选自二元crrna:tracrrna组合和嵌合sgrna之一的分离的向导rna形成的核糖核蛋白(rnp)复合物。在一些实施方案中,将分离的crrna和tracrrna的长度修饰和/或化学修饰形式与纯化的cas9蛋白(例如,seqidno.:407-410)、分离的编码cas9蛋白的mrna(例如,seqidno.:413)或编码cas9蛋白的基因(例如,seqidno.:411和412)组合在表达载体中。在某些测定中,可以将分离的crrna和tracrrna的长度修饰和/或化学修饰形式引入从编码cas9基因的内源表达盒稳定表达cas9蛋白的细胞系中。在其它试验中,可以将crrna和tracrrna的长度修饰和/或化学修饰形式与cas9mrna或cas9蛋白组合的混合物引入到细胞中。
实施例1
本实施例说明化学修饰和截短的向导rna在体外cas9dna切割试验中起作用。
合成了crrna和tracrrna寡核苷酸,其相对于所示wt序列具有各种化学修饰和截短(表1)。
表1:hprt1靶dna被具有各种crrna和tracrrna对的cas9核酸内切酶切割的体外生物化学研究。
寡核苷酸序列以5'-3'示出。小写字母=rna,加下划线=2'-o-甲基rna,斜体字=2'-氟rna。指出了rna寡核苷酸的长度(碱基)。指出了通过琼脂糖凝胶电泳可视化的具有每个crrna:tracrrna对的重组cas9对dna靶的相对切割效率,其中“+++”表示完全切割,“++”和“+”表示中间水平的切割,并且“-”表示无切割。
crrna含有与合适的“ngg”pam位点相邻的人hprt1基因中的位点匹配的19个碱基前间区序列向导序列。将来自人hprt1基因的938个碱基对区域克隆到pcr-blunt载体(lifetechnologies)中。在用于cas9切割测定之前,通过用限制性核酸内切酶xmai(newenglandbiolabs)消化将质粒线性化。下文示出hprt1靶片段的序列。目标pam位点用粗体字表示,并且前间区序列向导序列结合位点加有下划线。
hprt1目标序列。seqidno.:27.gaatgttgtgataaaaggtgatgctcacctctcccacacccttttatagtttagggattgtatttccaaggtttctagactgagagcccttttcatctttgctcattgacactctgtacccattaatcctccttattagctccccttcaatggacacatgggtagtcagggtgcaggtctcagaactgtccttcaggttccaggtgatcaaccaagtgccttgtctgtagtgtcaactcattgctgccccttcctagtaatccccataatttagctctccatttcatagtctttccttgggtgtgttaaaagtgaccatggtacactcagcacggatgaaatgaaacagtgtttagaaacgtcagtcttctcttttgtaatgccctgtagtctctctgtatgttatatgtcacattttgtaattaacagcttgctggtgaaaagga
通过重组spycas9(newenglandbiolabs)测试crrna和tracrrna对的直接降解含有人hprt1基因的克隆片段的线性化质粒dna的能力。将crrna:tracrrna在150nm浓度的双链体缓冲液(30mmhepesph7.5、100mm乙酸钾)中退火。spycas9(15nm)与crrna:tracrrna以1:1摩尔比在37℃下预温育10分钟。随后,加入3nm的线性化目标质粒,并在37℃下温育1小时。反应产物在1%琼脂糖凝胶上在125v下分离1小时。根据制造商的方案,通过gelred(biotium)后染色使条带可视化。将凝胶在uv透照仪上成像,结果总结于上表1中。
天然野生型(wt)crisprrna在5’末端具有19-20个碱基的前间区序列结构域(向导,其与靶核酸结合)并且在3’末端具有22个碱基的结构域,其与tracrrna结合。因此,wtcrrna的长度是41-42个碱基。wttracrrna的长度是89个碱基。我们观察到wt型crrna:tracrrna对支持目标dna的完全切割(cr/tracrrna对2d)。我们还观察到,与67个碱基的tracrrna配对的具有35个碱基的crrna(19个碱基的前间区序列和16个碱基的tracrrna结合结构域)的试剂的截短形式支持目标rna的完全切割(cr/tracrrna对1a)。crrnatracrrna结合区在3’末端被截短6个碱基(seqidno.:1)。tracrrna在两端都被截短(seqidno.:2)。短crrna与长tracrrna的成对组合显示出切割,长crrna与短tracrrna(对2a)也是如此。这些发现是重要的,因为它允许使用较短的rna组分来引导cas9靶识别和切割。相比于较长的rna寡核苷酸,较短的rna寡核苷酸对于化学合成来说较廉价且难度较小,需要较少的纯化且提供较高的产率。
缺失天然crrna和tracrrna的一些元件(图1)以制备功能性sgrna(图2)。然而,在sgrna中将crrna结合于tracrrna的双链核酸量限于11个碱基对,这对生物盐条件下的双链体形成来说通常太短。由于单分子发夹结构,复合物以sgrna形式是稳定的,但该序列分成2个rna将是不稳定的。因此,不清楚需要什么长度的双链结构域来制备最小但具功能性的2分子(2部分)crispr复合物,或者该复合物是否将用于引导cas9进行靶切割。本实施例表明,对于具有少至15个碱基,配对的碱基允许2部分crrna:tracrrna复合物的功能,其有效地引导cas9核酸酶针对与crrna前间区序列结构域(seqidno.:1和2)互补的靶的活性。
不耐受用2'omerna进行crrna的完全化学修饰(对3a和对5a)。此外,crrna的22个碱基tracrrna结合结构域的完全2'ome修饰不支持靶切割(对4a、对6a),并且前间区序列向导结构域的完全2'ome修饰不支持切割(对7a)。也不耐受用2'omerna进行tracrrna的完全化学修饰(对1b、1c和对2b、2c)。
重要的是,两种crisprrna物质的一些高度2'ome修饰形式都支持切割。对1k对于在3’末端具有29个2'ome残基的tracrrna(seqidno.:11)显示高切割活性。对1l对于5’末端的9个2'ome残基和3’末端的29个2'ome残基(seqidno.:13)显示高切割活性。因此,tracrrna的短形式中的67个rna残基中有38个可以在体外切割测定中转化为2'omerna(57%),而无活性损失。
对14a表明,在crrna的3’末端的11个碱基(22个碱基的tracrrna结合结构域的50%)可以用2'omerna修饰并支持靶切割(seqidno.:14)。当与修饰的tracrrna配对(对14l,seqidno.:13和14)时,修饰的crrna保持完全活性。crrna的5’末端的11个碱基(在向导前间区序列结构域中,碱基2-12)的修饰支持靶切割(对15a),并且该修饰当与修饰的tracrrna配对(对15l,seqidno.:13和15)时也具功能性。即使当与修饰的tracrrna配对(对16l,seqidno.:13和16)时,也可以将crrna的5’末端和3’末端的2'ome修饰组合(seqidno.:16),使得35个残基中的22个被修饰(63%)并且仍然支持切割(对16a)。
上述crrna:tracrrna对都使用2'omerna作为修饰剂。另外的研究表明,2'f修饰也被cas9耐受,并且能够切割目标dna。对9a在所有嘧啶碱基(seqidno.:23)上使用具有2'f修饰的crrna,并且该设计支持完全靶切割。crrna的完全2'f修饰也支持完全靶切割(对10a,seqidno.:24)。联合使用2'ome和2'f修饰可能允许crrna和tracrrna的完全修饰。本实施例中的研究利用体外生物化学分析。在哺乳动物基因编辑的背景下,性能可能改变,其中序列必须在细胞核环境中起作用。
实施例2
本实施例表明截短的tracrrna在哺乳动物细胞中起作用,通过spycas9核酸酶引导基因组编辑。
功能性cas9核酸酶和rna触发因子(单个sgrna或二元crrna:tracrrna对)必须存在于哺乳动物细胞核中,以进行crispr基因组编辑。表达cas9的大质粒载体的转染是无效的,并且增加了实验结果的可变性。为了准确评估crrna和tracrrna的长度和化学组成的变化在哺乳动物细胞中在不存在其它变量下的影响,构建了稳定表达spycas9的细胞系。
如下所述开发了具有稳定载体整合和在g418下选择的spycas9(人密码子优化)的组成型表达的hek293细胞系。将人优化spycas9连接到pcdna3.1表达载体(lifetechnologies)中,并使用lipofectamine2000(lifetechnologies)转染到hek293细胞中。使用新霉素使转染的细胞生长2天,然后置于选择性压力下。7天后,使用限制性稀释技术将细胞接种到单集落。筛选单克隆集落的cas9活性,并且将具有最高表达水平的克隆用于未来研究。使用液滴式数字pcr(ddpcr)确定cas9的单拷贝整合事件。使用抗cas9抗体的western印迹显示cas9蛋白的低但恒定的表达。该细胞系以下称为“hek-cas9”。
在随后的研究中使用hek-cas9细胞系。以反向转染方式,将抗hprt1crrna:tracrrna复合物与lipofectaminernaimax(lifetechnologies)混合并转染到hek-cas9细胞中。以96孔板格式用每孔40,000个细胞进行转染。在0.75μl脂质试剂中以30nm的最终浓度引入rna。将细胞在37℃下温育48小时。使用quickextract溶液(epicentre)分离基因组dna。利用kapahifidna聚合酶(roche)和靶向目标hprt区域的引物(hprt正向引物:aagaatgttgtgataaaaggtgatgct(seqidno.:28);hprt反向引物:acacatccatgggacttctgcctc(seqidno.:29))扩增基因组dna。将pcr产物在neb缓冲液2(newenglandbiolabs)中熔融并再退火,以允许异源双链体形成,接着用2单位的t7核酸内切酶1(t7ei;newenglandbiolabs)在37℃下消化1小时。消化产物在片段分析仪(advancedanalytics)上可视化。靶向dna的切割百分比计算为:切割产物的平均摩尔浓度/(切割产物的平均摩尔浓度+未切割条带的摩尔浓度)×100。
合成了tracrrna(表2),其在5’末端、3’末端、内部或其组合具有缺失。tracrrna与未修饰的截短的抗hprt1crrnaseqidno.:1(表1)复合,其在5’末端具有靶向hprt1的19个碱基的前间区序列结构域并且在3’末端具有16个碱基的tracrrna结合结构域。将配对的crrna:tracrrnarna寡核苷酸转染到hek-cas9细胞中并如上所述进行处理。通过比较使用t7ei错配核酸内切酶切割测定得到的hprt1基因的切割率与使用片段分析仪进行的产物的定量测量,来评估相对基因编辑活性。野生型酿酒酵母crrna:tracrrna复合物的描述在图1示出(其将crrnaseqidno.:46与tracrrnaseqidno.:18配对)。在该实施例中测试的tracrrna中缺失的相对位置在图3中以序列比对格式示出。
表2:在tracrrna中进行长度截短对哺乳动物细胞中cas9核酸内切酶的基因编辑效率的影响。
寡核苷酸序列以5'-3'示出。小写字母=rna。指出了rna寡核苷酸的长度(碱基)。指出了在截短研究中在5’末端、3’末端和内部(int)除去的rna残基数。每个物质的相对功能活性由t7ei异源双链体测定中的切割%指示。
本实施例表明,为了在哺乳动物细胞中进行基因编辑,tracrrna可以耐受5’末端和3’末端的显著缺失并保留完整功能性。从5’末端缺失18个碱基是良好耐受的。从5’末端缺失20个碱基导致活性降低,这可能是由于crrna的结合亲和力较低。如果使用tm增强修饰来稳定短的双链体形成区域,则这种缩短的长度或更短的长度可能具功能性。从3’末端缺失多达10个碱基是良好耐受的。额外的缺失导致活性损失。破坏发夹元件或发夹元件之间的间隔的内部缺失不具功能性。
总之,本实施例表明,将tracrrna从野生型(wt,seqidno.:18)的89个碱基长度截短到67个碱基长度(seqidno.:2)或截短到62个碱基长度(seqidno.:38)或截短到61个碱基长度(seqidno.:39)保持高功能活性。使用这种缩短的tracrrna相比于wt89个碱基rna将更廉价并且更容易通过化学方法制造。一些截短的物质(seqidno.:2、seqidno.:38和seqidno.:39)显示出比89个碱性wttracrrna增加的功能活性。因此,本发明的缩短的tracrrna除了成本较低且更容易通过化学方法制造之外,还显示出改进的活性。
实施例3
实施例1和2表明,比wt长度分别短42个和89个碱基的crrna:tracrrna复合物可以在哺乳动物基因编辑中显示更高的功能活性。本实施例显示了这些rna物质的长度的进一步优化。
合成了一系列crrna和tracrrna(表3),其具有所示的不同长度。在crrna的3’末端、tracrrna的5’末端和/或tracrrna的3’末端进行截短。crrna和tracrrna如表3中所示进行配对。crrna都采用了5’末端靶向hprt1的20个碱基的前间区序列结构域和可变长度3’末端(tracrrna结合结构域)。在该实施例中研究的crrna和tracrrna序列的比对在图4中示出,以阐明相对于每个功能结构域的截短位置。
将配对的crrna:tracrrnarna寡核苷酸转染到hek-cas9细胞中,并如实施例2中所述进行处理。通过比较使用t7ei错配核酸内切酶切割测定得到的hprt1基因的切割率与使用片段分析仪进行的产物的定量测量,来评估相对基因编辑活性。结果示于表3中。缺失的相对位置以序列比对格式示于图4中。
表3:在crrna和tracrrna中进行长度截短对哺乳动物细胞中cas9核酸内切酶的基因编辑效率的影响。
寡核苷酸序列以5'-3'示出。小写字母=rna。指出了rna寡核苷酸的长度(碱基)。每个crrna:tracrrna对的相对功能活性由t7ei异源双链体测定中的切割%指示。
所研究的所有化合物引导在hek-cas9细胞中的hprt1基因座处的crispr/cas编辑。效率在6%到57%之间广泛变化。最有效的crrna+tracrrna组合是36个碱基crrna(seqidno.:48)与67聚体tracrrna(seqidno.:2)。截短、优化的crrna:tracrrna复合物的示意图示于图5中。在这种情况下,crrna的tracrrna结合结构域从wt22个碱基序列(3’末端)截短到16个碱基。这与tracrrna的5’末端的crrna结合结构域杂交。tracrrna在5’末端截短18个碱基并且在3’末端截短4个碱基,产生具活性的67个碱基的产物。对于该对,在crrna的3’末端与tracrrna的5’末端杂交时形成钝端。其它形式也显示高活性,包括与70个碱基tracrrna(seqidno.:51)配对的42个碱基(wt)crrna(seqidno.:46)。
所测试的最短crrna的长度为34个碱基(seqidno.:49),并且相比于较长的变体通常表现出更低的活性。在该变体与tracrrna之间形成的较短的双链体结构域,与36个碱基crrna变体相比,具有降低的结合亲和力(tm),并且34个碱基复合物在37℃下较不稳定。使用增加结合亲和力(tm)的化学修饰,例如2'omerna、2'frna或lna残基,将增加该短双链体结构域的稳定性,并且将导致改进的活性,从而允许使用这种设计的非常短的crrna。广泛使用tm增强修饰将允许在crrna中使用更短的tracrrna结合结构域,例如13个碱基、或12个碱基、或11个碱基、或10个碱基、或9个碱基、或8个碱基或更短,这取决于所用的修饰残基的种类和数量。
实施例4
实施例1、2和3表明,比分别为42个和89个碱基的wt长度短的crrna:tracrrna复合物可以在哺乳动物基因编辑中显示更高的功能活性。在这些实施例中,所有的截短都是在rna的通用结构域中进行的。本实施例测试了截短对向导crrna的靶特异性前间区序列结构域的影响。
合成了一系列crrna(表4),其具有如所示的20、19、18或17个碱基长度的前间区序列结构域。使用3’末端的16聚体通用tracrrna结合序列,在crrna的5’末端进行截短。将crrna与未修饰的67聚体tracrrna(seqidno.:2)进行配对。crrna靶向人hprt1基因的相同外显子中的4个不同位点。
将配对的crrna:tracrrnarna寡核苷酸转染到hek-cas9细胞中并如实施例2中所述进行处理。通过比较使用t7ei错配核酸内切酶切割测定得到的hprt1基因的切割率与使用片段分析仪进行的产物的定量测量,来评估相对基因编辑活性。结果示于表4中。
表4:在crrna的5'-前间区序列结构域中进行长度截短对哺乳动物细胞中cas9核酸内切酶的基因编辑效率的影响。
寡核苷酸序列以5'-3'示出。小写字母=rna。靶特异性前间区序列结构域加有下划线并且指出了长度(碱基)。每个物质的相对功能活性由t7ei异源双链体测定中的切割%指示。
在所研究的4个位点中,一个(位点38087)对于所有4种crrna都显示出高活性,随着前间区序列结构域缩短,活性没有变化。位点38285对于20个碱基和19个碱基的前间区序列crrna(seqidno.:48和1)显示类似功效,对于18个碱基形式(seqidno.:54)略有降低,并且对于17个碱基形式(seqidno.:55)大幅降低。位点38094对于20个碱基和19个碱基的前间区序列crrna(seqidno.:64和65)显示类似功效,对于18个碱基形式(seqidno.:66)中等降低,并且对于17个碱基形式(seqidno.:67)无活性。位点38358对于20个碱基形式(seqidno.:60)显示出良好活性,对于19个碱基形式(seqidno.:61)的活性较低,对于18个碱基形式(seqidno.:62)的活性更低,并且对于17个碱基形式(seqidno.:63)无活性。
与野生型20个碱基结构域相比,使用缩短的17个碱基前间区序列向导结构域可以降低不期望的脱靶事件的发生(fu等,《自然:生物技术(naturebiotechnol.)》,32:279,2014)。我们观察到中靶功效以序列背景特异性方式改变,并且20个碱基和19个碱基前间区序列向导结构域是通常有效的,但是当使用18个碱基前间区序列结构域时活性开始降低,并且当使用17个碱基前间区序列结构域时活性显著降低。因此,为了维持所期望的中靶效率,在本文中使用20个和19个碱基靶特异性前间区序列向导结构域。前间区序列向导结构域的显著截短在许多情况下降低了cas9核酸内切酶对dna靶的中靶切割。使用增强tm的化学修饰(增加crrna的前间区序列靶特异性结构域与目标dna序列的结合亲和力)可以允许使用较短的序列,使得17个碱基的前间区序列向导结构域可显示与未修饰的20个碱基的前间区序列向导结构域类似的中靶功效。
实施例5
本实施例表明,截短的crrna:tracrrna复合物在多个位点显示改进的基因编辑活性。先前的实施例研究了截短的rna在人hrpt1基因中的单个位点作为哺乳动物细胞中crispr基因编辑的触发因子的功效。可能存在位点/序列特异性效应。本实施例表明,本发明的截短种类在人hprt1基因的外显子中的12个位点的性能得以改进。
合成了一系列crrna(表5),其具有对人hprt1基因中的12个位点具有特异性的长度为20个碱基的前间区序列结构域与在3’末端的16聚体通用tracrrna结合序列。将crrna与未修饰的67聚体tracrrna(seqidno.:2)配对。使用wt长度crrna:tracrrna复合物研究相同的12位点,其中crrna包含与3’末端的22聚体通用tracrrna结合序列配对的20个碱基的前间区序列向导结构域,其与wt89聚体tracrrna(seqidno.:18)复合。
将配对的crrna:tracrrnarna寡核苷酸转染到hek-cas9细胞中并如实施例2中所述进行处理。通过比较使用t7ei错配核酸内切酶切割测定得到的hprt1基因的切割率与使用片段分析仪进行的产物的定量测量,来评估相对基因编辑活性。结果示于表5中。
表5:在crrna和tracrrna中进行长度截短对哺乳动物细胞中cas9核酸内切酶的基因编辑效率的影响。
寡核苷酸序列以5'-3'示出。小写字母=rna。指出了rna寡核苷酸的长度(碱基)。每个crrna:tracrrna对的相对功能活性由t7ei异源双链体测定中的切割%指示。
在hek-cas9细胞中crispr介导的基因编辑的相对效率随序列背景而变化。然而,在所有情况下,较短的优化rna向导(36聚体crrna和67聚体tracrrna)相比于wtrna(42聚体crrna和89聚体tracrrna)显示更高的效率。使用本发明的缩短的优化向导rna相对于wtrna改进了靶向dna的cas9切割,从而提高了基因编辑速率。
实施例6
实施例1描述了在靶dna体外生物化学切割测定中与cas9一起起作用的化学修饰模式。本实施例证实了化学修饰的tracrrna在哺乳动物细胞中起作用,引导通过spycas9核酸酶进行基因组编辑。体外和体内使用之间,最佳修饰模式不同。
合成了一系列tracrrna(表6),其具有多种化学修饰,包括:核糖修饰2'omerna和lna;末端修饰基团丙二醇间隔基和萘基-偶氮修饰剂(n,n-二乙基-4-(4-硝基萘-1-基偶氮)-苯胺,或“zen”);和具有硫代磷酸酯修饰的所选核苷酸间的键。关于萘基-偶氮修饰剂的结构以及萘基-偶氮修饰剂和丙二醇修饰剂用作端基来阻止核酸外切酶攻击的用途,参见:lennox等,《分子疗法:核酸(moleculartherapynucleicacids)》,2:e1172013。表6中列出的tracrrna与在5’末端具有靶向hprt1的19个碱基的前间区序列结构域和在3’末端具有16个碱基的tracrrna结合结构域的未修饰的截短的抗hprt1crrnaseqidno.:1(表1)复合。将配对的crrna:tracrrnarna寡核苷酸转染到hek-cas9细胞中并如上所述进行处理。通过比较使用t7ei错配核酸内切酶切割测定得到的hprt1基因的切割率与使用片段分析仪进行的产物的定量测量,来评估相对基因编辑活性。
表6:哺乳动物细胞中的tracrrna寡核苷酸修饰模式的优化。
寡核苷酸序列以5'-3'示出。大写字母=dna;小写字母=rna;加下划线=2'-o-甲基rna;斜体字=2'-氟rna;+a、+c、+t、+g=lna;c3=c3间隔基(丙二醇修饰剂);*=硫代磷酸酯核苷酸间的键;zen=萘基-偶氮修饰剂;inv-dt=反向dt。每个物质的相对功能活性由t7ei异源双链体测定中的切割%指示。
由于存在降解未修饰的寡核苷酸的核酸外切酶和核酸内切酶,修饰通常是合成核酸在细胞内环境中充分起作用所必需的。已经描述了赋予寡核苷酸以核酸酶抗性的广泛多种的修饰。对于给定应用充分起作用的所用修饰的精确组合和顺序可以随着序列背景和生物功能所需的蛋白质相互作用的性质而变化。关于反义寡核苷酸(其与rna酶h1相互作用)和sirna(其与dicer、ago2和其它蛋白质相互作用)的化学修饰已经进行了大量先前研究。预期化学修饰会改进crisprcrrna:tracrrna复合物的功能。然而,不可能预测什么样的修饰和/或修饰模式将与合成rna与cas9的功能复合相容。本发明定义了tracrrna的最小、中等和大量化学修饰模式,其保持高水平的功能以在哺乳动物细胞中引导cas9介导的基因编辑。
表6中的结果表明,在tracrrna的整个5'和3’末端结构域内耐受大量修饰。tracrrna的内部结构域的修饰显示出降低的活性,这可能是由于折叠的rna的结构改变和/或通过2'ome修饰使得蛋白质接触点被关键rna残基的2'-oh阻断。例如,化合物seqidno.:100具有用2'omerna修饰的39/67残基(58%),并且与未修饰的序列相比保持完全活性。seqidno.:134具有用2'omerna修饰的46/67残基(69%),并且与未修饰的序列相比保持接近完全活性(图6)。seqidno.:134是tracrrna的截短的67聚体变体。使用seqidno.:134作为模型,耐受用2'omerna修饰5'结构域中的11个连续残基而无活性损失。耐受用2'omerna修饰3'结构域中的35个连续残基而无活性损失。值得注意的是,存在于3'结构域中的两个发夹结构对于功能是必需的,因为这些特征中的任一个的缺失导致活性损失(实施例2、图3),但是这两个结构域都可以用2'omerna完全修饰而不损害功能。注意,seqidno.:134和100在5’末端和3’末端都具有硫代磷酸酯(ps)修饰的核苷酸间的键,这为核酸外切酶攻击提供了额外的保护。
鉴定了当被修饰时导致活性大幅降低或完全损失的特定残基。使用67个碱基tracrrna(例如,seqidno.:134)作为参考,从序列的5’末端开始,在残基u12、a15、g26、u27、g30、u31和u32处用2'omerna取代天然rna导致活性大量损失(图6)。此外鉴定了当被修饰时导致活性较小但显著降低的特定残基。使用67个碱基tracrrna(例如,seqidno.:134)作为参考,从序列的5’末端开始,在残基u13、u18、c23、u24和c28处用2'omerna取代天然rna导致活性降低(图6)。本研究使用2'omerna进行。在这些位置使用其它修饰如2'f、lna、dna等可以更好地耐受。seqidno.:134中的未修饰rna的中心21残基结构域用2'-frna完全修饰(seqidno.:141)或部分修饰(seqidno.:142和143)。这些变体不具功能性。seqidno.:134中的未修饰rna的中心21残基结构域用dna完全修饰(seqidno.:138)或部分修饰(seqidno.:139和140)。这些变体不具功能性。这个结构域中的孤立残基的修饰可能会起作用,但是在该结构域中连续的大段(block)修饰降低了tracrrna的活性。
为了进一步研究可以在tracrrna的中心结构域内使用2'omerna修饰哪些单独残基,进行了单个碱基修饰2'omerna‘步行’(seqidno.:144-162)。在本系列中,作为残基a14、a19、a20、g21、g22、a25和c29的修饰显示没有活性损失并且是修饰的候选者。
反义寡核苷酸通常使用完全ps修饰制得,其中每个核苷酸间的键是硫代磷酸酯修饰的。这种大量修饰水平是可能的,因为当形成功能性底物/酶复合物时,蛋白质效应分子rnaseh1(其介导aso引导的mrna降解)耐受aso中的ps修饰。另一方面,sirna不能耐受完全ps修饰;大量ps修饰破坏与效应蛋白ago2(其介导sirna引导的mrna降解)的有效的相互作用。内部rna环中tracrrna的大量ps修饰破坏了与cas9的功能相互作用(seqidno.:133;29个ps修饰)。可以进行有限的ps末端修饰而不损失活性(seqidno.:98和101;每一端有2-3个pp键)。中心结构域中可耐受较少量ps修饰。特别地,rna酶切割映射(其中使用一系列血清或细胞提取稀释液中的tracrrna的温育来找到对rna酶攻击最敏感的位点)可用于鉴定临界位点,其中仅一个或几个键的ps修饰可以稳定rna而不破坏功能。
存在其中ps修饰有助于化学毒性的应用。在这种情况下,需要使用其它方法来阻断核酸外切酶攻击。选项包括末端修饰剂,例如反向dt或脱碱基团如dspacer、c3间隔基(丙二醇)、zen(萘基-偶氮修饰剂)等。布置这种末端修饰基团可以消除末端ps核苷酸间的键的需要。
实施例7
实施例1描述了在靶dna体外生物化学切割测定中与cas9一起起作用的化学修饰模式。本实施例证实了化学修饰的crrna在哺乳动物细胞中起作用,引导spycas9核酸酶进行基因组编辑。体外和体内使用之间,最佳修饰模式不同。
合成了一系列crrna(表7),其具有多种化学修饰,包括:核糖修饰2'omerna、2'f和lna;末端修饰基团丙二醇间隔基和萘基-偶氮修饰剂(n,n-二乙基-4-(4-硝基萘-1-基偶氮)-苯胺或“zen”),和反向dt残基;和具有硫代磷酸酯修饰的所选核苷酸间的键。关于萘基-偶氮修饰剂的结构以及萘基-偶氮修饰剂和丙二醇修饰剂用作端基来阻止核酸外切酶攻击的用途,参见:lennox等,《分子疗法:核酸(moleculartherapynucleicacids)》,2:e1172013。crrna具有5’末端的靶向hprt1的19个碱基前间区序列结构域(seqidno.:1、9、10、14-16、22-24、163-173)或具有靶向相同位点的20个碱基前间区序列结构域(seqidno.:48、174-237)与3’末端的16个碱基tracrrna结合结构域。表7中列出的crrna与未修饰的截短(67个碱基)tracrrnaseqidno.:2(表1)或化学修饰的截短(67个碱基)tracrrnaseqidno.:100(表6)复合。使用两个tracrrna能够确定化学修饰的crrna的功能在与修饰的tracrrna配对时是否会改变。将配对的crrna:tracrrnarna寡核苷酸转染到hek-cas9细胞中并如前所述进行处理。通过比较使用t7ei错配核酸内切酶切割测定得到的hprt1基因的切割率与使用片段分析仪进行的产物的定量测量,来评估相对基因编辑活性。
表7:哺乳动物细胞中的crrna寡核苷酸修饰模式的优化。
寡核苷酸序列以5'-3'示出。大写字母=dna;小写字母=rna;加下划线=2'-o-甲基rna;斜体字=2'-氟rna;+a、+c、+t、+g=lna;c3=c3间隔基(丙二醇修饰剂);*=硫代磷酸酯核苷酸间的键;zen=萘基-偶氮修饰剂;invt=反向dt。当所示crrna与所示tracrrna配对时,每个物质的相对功能活性由t7ei异源双链体测定中的切割%指示。nd=未测定。
由于存在降解未修饰的寡核苷酸的核酸外切酶和核酸内切酶,合成核酸在细胞内环境中充分起作用通常需要某种化学修饰。已经描述了赋予寡核苷酸以核酸酶抗性的广泛多种修饰。对于给定应用充分起作用的所用修饰的精确组合和顺序可以随着序列背景和生物功能所需的蛋白质相互作用的性质而变化。关于反义寡核苷酸(其与rna酶h1相互作用)和sirna(其与dicer、ago2和其它蛋白质相互作用)的化学修饰已经进行了大量先前研究。预期化学修饰会改进crisprcrrna:tracrrna复合物的功能。然而,不可能预测什么样的修饰和/或修饰模式将与rna和cas9以功能方式进行的结合相容。本发明定义了用于在哺乳动物细胞中保持高水平功能以引导cas9介导的基因编辑的crrna的最小、中等和大量化学修饰模式。实施例7中的研究是靶向人hprt1基因中的单一位点进行的。应注意,表现良好的crrna的20个碱基5’末端前间区序列向导结构域的修饰模式可能随序列背景而变化。然而,当靶向不同位点时,当相邻前间区序列结构域的序列改变时,很可能会影响如本文所定义的表现良好的3’末端tracrrna结合结构域的修饰模式,因此这里显示的3'结构域修饰模式将是“通用的”。
表7中的结果表明,在crrna的整个5'和3’末端内耐受大量修饰。crrna的内部结构域内的某些所选位置的修饰导致活性降低或活性完全阻断,这可能是由于折叠rna的结构改变和/或通过2'ome修饰使得蛋白质接触点被关键rna残基的2'-oh阻断。例如,化合物seqidno.:204具有用2'omerna修饰的21/36残基(58%),并且与未修饰序列相比保持完全活性。化合物seqidno.:239具有用2'omerna修饰的30/36残基(83%),并且与未修饰的序列相比保持完全活性。这两种化合物在5'和3’末端也都具有3个硫代磷酸酯(ps)修饰的核苷酸间的键,这为核酸外切酶攻击提供额外的保护。相比之下,seqidno.:165仅具有用2'omerna修饰的4/36残基(11%),但完全失去活性。
大段序列对crrna的5’末端和3’末端的2'ome修饰是耐受的,然而在分子的中心部分的某些残基的修饰导致失活。为了进一步研究可以在crrna的中心结构域内使用2'omerna修饰哪些单独残基,进行了单个碱基修饰2'omerna‘步行’(seqidno.:272-286)。鉴定了导致活性大量降低或完全损失的特定残基(crrna内的位置)。使用36个基因的crrnaseqidno.:239作为模型并从序列的5’末端开始编号,用2'omerna取代残基u15和u16的天然rna导致活性显著损失,并且取代残基u19的天然rna导致活性中度损失(图7)。这3个位点位于靶特异性前间区序列向导结构域内,因此序列随靶而变化(残基15、16和19,图7)。在某些序列背景中,这些位点可能会耐受修饰。在通用tracrrna结合结构域(残基21-36)内,用2'omerna取代残基u22、u23和u24的天然rna导致活性大量损失。鉴于该结构域不会随着序列背景而改变,这些位点的修饰耐受性可能将不会随着目标序列变化而改变。在实施例10中更详细地研究了20个碱基靶特异性前间区序列向导结构域中的修饰的序列特异性作用。
通常制备具有完全ps修饰的反义寡核苷酸,其中每个核苷酸间的键是硫代磷酸酯修饰的。这种大量修饰水平是可能的,因为当形成功能性底物/酶复合物时,蛋白质效应分子rna酶h1耐受aso中的ps修饰。另一方面,sirna不能耐受完全ps修饰;大量ps修饰破坏了与效应蛋白ago2的生产性相互作用。可以在不损失活性的情况下进行crrna的有限ps末端修饰(seqidno.:177、178、239等,每端具有3个ps键)。末端修饰是需要的,因为这增加了对核酸外切酶攻击的额外保护。所选内部位点的ps修饰也可以被耐受,并且可能为核酸内切酶攻击提供额外的保护。使用seqidno.:264作为碱基修饰模式,在tracrrna结合结构域(seqidno.:265)中,在前间区序列向导结构域的3’末端(种子区域)(seqidno.:266)中或两个区域(seqidno.:267)中对内部的键进行ps修饰。ps修饰水平增加导致功能活性降低,其中seqidno.:267具有的活性是较少修饰的seqidno.:264变体的约50%。seqidno.:267中的35个核苷酸间的键中有21个被修饰并且对核酸酶暴露将是稳定的。在需要暴露于高核酸酶环境的情况下(例如用于研究或治疗适应症的直接iv施药),这种高度修饰的变体相比于较少修饰的变体(其将更快地降解)实际上可以显示更高的活性。
存在其中ps修饰促进化学毒性的实验设置。在这种情况下,需要使用其它方法来阻断核酸外切酶攻击。crrna可以具有置于5’末端和3’末端中的任一个或两个上的c3隔离基(丙二醇修饰剂)或zen(萘基-偶氮修饰剂),以阻断核酸外切酶攻击,从而避免了ps修饰的需要。该策略可用于消除ps封端修饰(参见seqidno.:179-186)。该策略可用于降低更高度修饰的crrna变体的ps含量。seqidno.:271具有与seqidno.:267相同模式的内部前间区序列结构域和ps修饰的racrrna结合域,但仅使用15个ps核苷酸间的键(而不是21个),并且显示改进的活性。因此,非碱基封端与内部ps修饰的组合可用于提高核酸酶稳定性,同时保持高活性。
实施例8
以下实施例证明本发明的经修饰的crisprcrrna和tracrrna的改善的效能。实施例2-7采用将crrna:tracrrna复合物以30nm浓度转染到人hek-cas9细胞中。实验测试先前表明,该剂量代表了剂量反应曲线的上限,使得使用较高剂量的rna不能改善基因编辑效率,但使用较低剂量导致较低的基因编辑效率。这些测量使用未修饰的rna进行。本实施例重新研究了本发明的新的优化的化学修饰的rna与未修饰的rna相比的剂量反应,并且证明化学修饰(即,核酸酶稳定化)导致更有效的化合物,其可以以较低剂量使用。
实施例5表明,本发明的截短向导rna在人hprt1基因中的12个位点的表现优于wtrna。选择这些位点中的四个(38087、38231、38133和38285),用于本实施例中未修饰rna与修饰rna的比较。未修饰的crrna与未修饰的tracrrna(seqidno.:2)以1:1摩尔比配对。未修饰的crrna与修饰的tracrrna(seqidno.:100)以1:1摩尔比配对。修饰的crrna与修饰的tracrrna(seqidno.:100)以1:1摩尔比配对。序列示于表8中。如前所述在30nm、10nm和3nm浓度下将rna转染到hek-cas9细胞中。将细胞在37℃下温育48小时,然后处理dna,并研究基因编辑活性的证据,其比较了t7ei错配核酸内切酶测定中的hprt1基因座处的切割率与如先前所述使用片段分析仪进行的产物的定量测量。结果示于表8中。
表8:修饰对未修饰的crrna:tracrrna复合物在哺乳动物细胞中引导cas9介导基因编辑的效能增加。
寡核苷酸序列以5'-3'示出。小写字母=rna;加下划线=2'-o-甲基rna;*=硫代磷酸酯核苷酸间的键。未修饰的crrna=un-cr。未修饰的tracrrna=un-tr。修饰的crrna=mod-cr。修饰的tracrrna=mod-tr。对于所研究的每个剂量,每个种类的相对功能活性由t7ei异源双链体测定中的切割%指示。
一般来说,当rna以高剂量转染时(其中rna过量存在),crrna和tracrrna的修饰对基因编辑效率的影响很小。在较低剂量下,改进的试剂显示改进的效能,并且在某些情况下显示显著改进的效能。改进程度随位点而变化。非常有效的位点38087显示了所测试的所有crrna/tracrrna变体在30nm和10nm剂量下的高效率基因编辑,但是在3nm下使用修饰的tracrrna(与任一个crrna)显示改进的活性。低效能位点,例如38231,即使在使用修饰rna测试的最高剂量(30nm)下也显示改进的基因编辑效率。单独的tracrrna修饰显示益处,但是当crrna和tracrrna都被修饰时,实现了最大的益处。图8示出了对hprt1位点38285具特异性的一个有效的修饰crrna(seqidno.:178)与修饰tracrrna(seqidno.:100)配对的示意图。图9示出了也对hprt1位点38285具特异性的高功能性crrna(seqidno.:239)与修饰tracrrna(seqidno.:134)配对的更高度修饰对的示意图。
本实施例将crrna:tracrrna复合物转染到hek-cas9细胞中,其中cas9蛋白被组成型表达。因此,转染的rna可以立即结合cas9蛋白,最小化细胞质中的核酸酶降解风险。可以预期,在转染的rna必须在暴露于细胞核酸酶后保持活性(survive)且同时产生cas9蛋白的情况下,crrna和/或tracrrna的化学修饰的益处将更大,如当使用其中cas9mrna或cas9表达载体与靶向rna共转染以使得cas9尚未在细胞中表达的方案所发生的那样。使用高度修饰的rna的益处对于体内应用(例如医学治疗剂)将是最大的,其中rna可能暴露于血清中存在的两种核酸酶(iv施药后)和细胞胞质核酸酶。
实施例9
实施例2-8表明截短和/或化学修饰的crisprcrrna和/或tracrrna在组成型表达cas9的哺乳动物细胞中触发cas9介导的基因组编辑的活性。本实施例表明本发明的截短的经修饰的rna组合物可以结合cas9蛋白,并且该复合物可以转染到人细胞中,并且此外核糖核蛋白(rnp)复合物的转染足以触发高效基因组编辑。
在本实施例中使用了对人hprt1位点38285具特异性的试剂。未修饰的crrna与未修饰的tracrrna以1:1摩尔比配对。未修饰的crrna与修饰的tracrrna以1:1摩尔比配对。修饰的crrna与修饰的tracrrna以1:1摩尔比配对。序列示于表9中。如上所述将rna转染到未修饰的hek293细胞中,不同之处在于使用增加量的rnaimax脂质转染试剂(1.2μl,相比于在hek-cas9细胞中以96孔格式对于仅30nmrna转染的每100μl转染使用的0.75μl量有所增加),以10nm浓度使用cas9蛋白(cariboubiosciences)与crrna:tracrrna的1:1复合物。将细胞在37℃温育48小时,然后处理dna并研究基因编辑活性的证据,其比较了t7ei错配核酸内切酶测定中的hprt1基因座处的切割率与如先前所述使用片段分析仪进行的产物的定量测量。结果示于表9中。
表9:修饰vs.未修饰的crrna:tracrrna复合物在哺乳动物细胞中引导cas9介导基因编辑的效能增加。
寡核苷酸序列以5'-3'示出。小写字母=rna;加下划线=2'-o-甲基rna;*=硫代磷酸酯核苷酸间的键。未修饰的crrna=un-cr。未修饰的tracrrna=un-tr。修饰的crrna=mod-cr。修饰的tracrrna=mod-tr。对于所研究的每个剂量,每个复合物的相对功能活性由t7ei异源双链体测定中的切割%指示。
所有3种crisprrna复合物在哺乳动物基因组编辑的rnp转染方案中都表现良好。在rnp方案中,未修饰的crrna+未修饰的tracrrna对(seqidno.:48和2)和未修饰的crrna+修饰的tracrrna对(seqidno.:48和100)在10nm剂量下比在hek-cas9方案中的表现好2.5倍,这与在细胞质或细胞核中cas9蛋白的转染与最终复合之间遭受降解的较少修饰的rna一致。因此,未修饰的rna需要较高的剂量,并且在某些情况下,未修饰的rna很可能不能引导任何基因组编辑活性。另一方面,修饰的crrna+修饰的tracrrna(seqidno.:178和100)在两种方案中都具有高效率。
本发明的修饰的截短的crisprrna良好作用于直接cas9rnp转染方法。
实施例10
实施例6和7中进行的化学修饰优化研究已研究了与具有各种修饰模式的tracrrna配对的具有各种修饰模式的crrna的活性。tracrrna是通用的,并且在所有目标位点处使用相同的序列。预期tracrrna的各种修饰模式的性能在不同的目标位点之间将是相似的。然而,crrna在不同的目标位点之间的序列改变。在实施例7和8中测试的优化形式中,crrna的5'-20个碱基具靶特异性(即“前间区序列结构域”)并且3'-16个碱基是通用的(即“tracrrna结合结构域”)。像tracrrna一样,预期crrna的通用16个碱基3'结构域中的各种修饰模式的性能在所有目标位点处将是相似的。然而,可能的是,不同修饰模式的性能可能受到5'-20个碱基靶特异性结构域中存在的序列背景的影响。
已充分确定的是,小干扰rna(sirna)的有效修饰模式受到序列背景的影响(behlke,《寡核苷酸(oligonucleotides)》,18:305-320,2008)。对于sirna,某些“有限修饰”模式可以应用于所有位点,而对于“大量修饰”,不可能预测哪个模式对于给定的序列将具功能性,并且经验测试是必要的。本实施例研究了序列背景对crrna的影响,在不同位点处测试了5'-20个碱基靶特异性结构域内的不同修饰模式。
实施例6和7中的修饰研究在人hprt1基因中使用单个crrnapam位点。本研究检查了人hprt1基因中的12个位点,包括先前检查的位点,其比较了不同修饰模式的功能性能,并建立了可以在所有位点以良好结果使用的单一修饰模式。关于这12个位点的其它研究,参见实施例5。
合成了一系列crrna(表10),其具有对人hprt1基因中的12个位点具特异性的长度为20个碱基的前间区序列结构域与3’末端的16聚体通用tracrrna结合序列。使用多种化学修饰制备了crrna,包括:核糖修饰2'omerna、末端修饰基团丙二醇间隔基和萘基-偶氮修饰剂(n,n-二乙基-4-(4-硝基萘-1-基偶氮)-苯胺或“zen”)、反向dt残基;和具有硫代磷酸酯修饰的选择核苷酸间的键。所采用的不同修饰模式的示意图示于图10中。
将crrna与高度修饰的67聚体tracrrna(seqidno.:100)配对。将配对的crrna:tracrrnarna寡核苷酸转染到hek-cas9细胞中并按实施例2中所述进行处理。通过比较使用t7ei错配核酸内切酶切割测定得到的hprt1基因的切割率与使用片段分析仪进行的产物的定量测量,来评估相对基因编辑活性。结果示于表10和图11中。
表10:在哺乳动物细胞中12个目标位点的crrna寡核苷酸修饰模式的优化。
寡核苷酸序列以5'-3'示出。小写字母=rna;加下划线=2'-o-甲基rna;c3=c3间隔基(丙二醇修饰剂);*=硫代磷酸酯核苷酸间的键;zen=萘基-偶氮修饰剂。当所示crrna与所示tracrrna在人hrpt1中的12个位点中的每一个处配对时,每个物质的相对功能活性由t7ei异源双链体测定中的切割%指示。
修饰的crrna在16碱基3’末端结构域中采用固定修饰模式,其是通用的并结合tracrrna。在靶特异性的5’末端结构域中测试/比较不同的修饰模式(即序列随目标位点而变化)。测试组包含具有在5’末端开始并朝向3’末端行进的0、3、4、6、8、10、12、13或14个连续2'omerna残基的变体。修饰模式避免了先前证明降低crrna功能性能的位置(实施例7)。也测试了仅非碱基修饰剂端基(c3间隔基或zen)的使用(无额外修饰)。当调查中比较所有12个位点的功能活性时,所测试的所有位点在5’末端的0-10个rna残基被2'omerna残基代替时显示完全活性。12个位点中仅一个显示出12个残基被修饰时活性略有降低,但是当13个残基被修饰时3/12位点显示活性降低,并且当14个残基被修饰时4/12位点显示活性降低。末端修饰剂(c3、zen)在所有位点都显示出完全活性。
在所测试的所有位点显示完全活性的最高水平的crrna修饰包括修饰模式6和7(图10)。这分别代表crrna中的碱基的61%和67%用2'omerna修饰。
n*n*n*nnnnnnnnnnnnnnnnnguuuuagagcuau*g*c*u修饰模式6
(seqidno.:434)
n*n*n*nnnnnnnnnnnnnnnnnguuuuagagcuau*g*c*u修饰模式7
(seqidno.:435)
本实施例中的数据还表明,个别位点可以在更高的水平上修饰并保留效能。例如,使用修饰模式8研究的12个位点中的8个显示完全活性,其中72%的残基被修饰。此外,实施例7表明,hprt1(seqidno.:239)中的靶向位点38285的crrna具有完全活性,并且30/36残基被修饰(83%,仅剩余6个未修饰的rna残基)。碱基修饰模式如修饰模式6或修饰模式7可以用作研究的起点,以凭经验确定在活性损失之前特定序列可以被修饰的程度。图12显示了修饰模式6crrna与高度修饰的tracrrna(seqidno.:134)配对的示意图。
实施例11
此处实施例使用来自化脓性链球菌的cas9核酸内切酶。s.py.cas9(spycas9)的天然氨基酸序列如下所示(seqidno.:407)。
天然cas9dna序列经密码子优化以在大肠杆菌中表达,并且具有为哺乳动物核定位(核酸酶定位信号)并辅助蛋白质纯化(his标签)而添加的元件。显示了重组蛋白的最终氨基酸序列(seqidno.:408)。显示了用于在大肠杆菌中表达重组蛋白的dna序列(seqidno.:409)。
天然cas9dna序列经密码子优化以在人细胞中表达,并具有为抗体识别(v5表位)和哺乳动物核定位(核酸酶定位信号,nls)而添加的元件。显示了最终的氨基酸序列(seqidno.:410),并且dna序列如下(seqidno411)。
天然的s.pycas9dna序列被密码子优化以在人细胞中表达并组装为t7rna聚合酶表达盒(seqidno.:412)。该序列包含t7rna聚合酶启动子、v5表位标签、核定位信号、密码子优化的cas9序列、第二核定位信号和具有多聚腺苷酸化信号的bgh(牛生长激素)基因3'-utr元件。显示了由该表达盒制备的mrna的序列(seqidno.:413)。
s.py.cas9氨基酸序列(seqidno.:407)。
mdkkysigldigtnsvgwavitdeykvpskkfkvlgntdrhsikknligallfdsgetaeatrlkrtarrrytrrknricylqeifsnemakvddsffhrleesflveedkkherhpifgnivdevayhekyptiyhlrkklvdstdkadlrliylalahmikfrghfliegdlnpdnsdvdklfiqlvqtynqlfeenpinasgvdakailsarlsksrrlenliaqlpgekknglfgnlialslgltpnfksnfdlaedaklqlskdtydddldnllaqigdqyadlflaaknlsdaillsdilrvnteitkaplsasmikrydehhqdltllkalvrqqlpekykeiffdqskngyagyidggasqeefykfikpilekmdgteellvklnredllrkqrtfdngsiphqihlgelhilrrqedfypflkdnrekiekiltfripyyvgplargnsrfawmtrkseetitpwnfeevvdkgasaqsfiermtnfdknlpnekvlpkhsllyeyftvyneltkvkyvtegmrkpaflsgeqkkaivdllfktnrkvtvkqlkedyfkkiecfdsveisgvedrfnaslgtyhdllkiikdkdfldneenediledivltltlfedremieerlktyahlfddkvmkqlkrrrytgwgrlsrklingirdkqsgktildflksdgfanrnfmqlihddsltfkediqkaqvsgqgdslhehianlagspaikkgilqtvkvvdelvkvmgrhkpeniviemarenqttqkgqknsrermkrieegikelgsqilkehpventqlqneklylyylqngrdmyvdqeldinrlsdydvdhivpqsflkddsidnkvltrsdknrgksdnvpseevvkkmknywrqllnaklitqrkfdnltkaergglseldkagfikrqlvetrqitkhvaqildsrmntkydendklirevkvitlksklvsdfrkdfqfykvreinnyhhahdaylnavvgtalikkypklesefvygdykvydvrkmiakseqeigkatakyffysnimnffkteitlangeirkrplietngetgeivwdkgrdfatvrkvlsmpqvnivkktevqtggfskesilpkrnsdkliarkkdwdpkkyggfdsptvaysvlvvakvekgkskklksvkellgitimerssfeknpidfleakgykevkkdliiklpkyslfelengrkrmlasagelqkgnelalpskyvnflylashyeklkgspedneqkqlfveqhkhyldeiieqisefskrviladanldkvlsaynkhrdkpireqaeniihlftltnlgapaafkyfdttidrkrytstkevldatlihqsitglyetridlsqlggd
含有3个nls序列和纯化his标签的从经密码子优化以在大肠杆菌中表达的dna表达的s.pycas9氨基酸序列(seqidno.:408)。
mgssapkkkrkvgihgvpaamdkkysigldigtnsvgwavitdeykvpskkfkvlgntdrhsikknligallfdsgetaeatrlkrtarrrytrrknricylqeifsnemakvddsffhrleesflveedkkherhpifgnivdevayhekyptiyhlrkklvdstdkadlrliylalahmikfrghfliegdlnpdnsdvdklfiqlvqtynqlfeenpinasgvdakailsarlsksrrlenliaqlpgekknglfgnlialslgltpnfksnfdlaedaklqlskdtydddldnllaqigdqyadlflaaknlsdaillsdilrvnteitkaplsasmikrydehhqdltllkalvrqqlpekykeiffdqskngyagyidggasqeefykfikpilekmdgteellvklnredllrkqrtfdngsiphqihlgelhailrrqedfypflkdnrekiekiltfripyyvgplargnsrfawmtrkseetitpwnfeevvdkgasaqsfiermtnfdknlpnekvlpkhsllyeyftvyneltkvkyvtegmrkpaflsgeqkkaivdllfktnrkvtvkqlkedyfkkiecfdsveisgvedrfnaslgtyhdllkiikdkdfldneenediledivltltlfedremieerlktyahlfddkvmkqlkrrrytgwgrlsrklingirdkqsgktildflksdgfanrnfmqlihddsltfkediqkaqvsgqgdslhehianlagspaikkgilqtvkvvdelvkvmgrhkpeniviemarenqttqkgqknsrermkrieegikelgsqilkehpventqlqneklylyylqngrdmyvdqeldinrlsdydvdhivpqsflkddsidnkvltrsdknrgksdnvpseevvkkmknywrqllnaklitqrkfdnltkaergglseldkagfikrqlvetrqitkhvaqildsrmntkydendklirevkvitlksklvsdfrkdfqfykvreinnyhhahdaylnavvgtalikkypklesefvygdykvydvrkmiakseqeigkatakyffysnimnffkteitlangeirkrplietngetgeivwdkgrdfatvrkvlsmpqvnivkktevqtggfskesilpkrnsdkliarkkdwdpkkyggfdsptvaysvlvvakvekgkskklksvkellgitimerssfeknpidfleakgykevkkdliiklpkyslfelengrkrmlasagelqkgnelalpskyvnflylashyeklkgspedneqkqlfveqhkhyldeiieqisefskrviladanldkvlsaynkhrdkpireqaeniihlftltnlgapaafkyfdttidrkrytstkevldatlihqsitglyetridlsqlggdaapkkkrkvdpkkkrkvaaalehhhhhh
含有3个nls序列和纯化his标签的经密码子优化以在大肠杆菌中表达的s.pycas9dna序列(seqidno.:409)。
atgggcagcagcgccccaaagaagaagcggaaggtcggtatccacggagtcccagcagccatggacaaaaagtactctattggcctggatatcgggaccaacagcgtcgggtgggctgttatcaccgacgagtataaagtaccttcgaaaaagttcaaagtgctgggcaacaccgatcgccattcaatcaaaaagaacttgattggtgcgctgttgtttgactccggggaaaccgccgaggcgactcgccttaaacgtacagcacgtcgccggtacactcggcgtaagaatcgcatttgctatttgcaggaaatctttagcaacgagatggcaaaagtcgatgactcgtttttccaccgcctcgaggaaagctttctggtggaggaagacaaaaagcatgagcgtcacccgatcttcggcaacattgtcgatgaagtagcgtatcatgaaaaatacccaaccatttaccacttacgcaaaaagctggtggacagcactgacaaagctgatttgcgccttatctatttagccctggcacatatgattaagtttcgtggtcacttcctgatcgaaggagacttaaatcccgacaacagtgatgttgataaattgtttattcagcttgtccaaacttacaatcaactgttcgaggaaaacccgatcaatgcctccggtgtggatgcaaaagccattttaagtgcacgccttagcaagtcccgtcgcttagaaaaccttatcgcgcagctgcccggcgagaaaaagaatggtttgtttgggaaccttattgccttgagcttaggcctcaccccgaatttcaaaagtaatttcgatcttgcagaagacgccaaattacaactgtcgaaggatacttatgatgacgatctcgataatctgttagcgcagattggtgaccaatacgccgatctttttctggcggctaaaaatctgagcgacgccatcttgctttcggatattctccgcgttaacaccgaaatcacgaaagcgcctcttagtgccagcatgattaaacgttatgatgaacaccaccaggacctgaccttactcaaagcgttggttcgccagcaactgccagagaagtacaaagaaatcttctttgatcagtcaaagaatggttatgccggctatattgacgggggtgcaagccaagaggaattctacaaatttatcaagcctattctggagaaaatggatggcaccgaagagttattggtgaagcttaaccgtgaagacctcctgcggaaacagcgcacattcgataatggttcgatcccacaccaaatccatttgggggagttacacgctattttgcgtcgccaggaagacttttaccctttcctgaaggataaccgggagaaaattgagaagatccttacctttcgtattccgtattacgtaggccccttagcacggggtaatagccgtttcgcgtggatgacacggaagtcggaagagacgatcaccccgtggaacttcgaagaggtagtcgacaagggcgcatcagcgcagtcttttattgaacgtatgacgaatttcgataaaaacttgcccaatgagaaggtgcttccgaaacattccttgttatatgaatattttacagtttacaacgagctgaccaaggttaaatacgtgacggaaggaatgcgcaagcccgcttttcttagcggtgagcaaaaaaaggcgatcgtcgacctgttattcaaaacgaatcgtaaggtgactgtaaagcaactcaaagaagattacttcaaaaagattgagtgcttcgacagcgtcgaaatctctggggtagaggatcggtttaacgcaagtttaggtacctaccatgacctgcttaaaatcattaaggataaagacttcttagataatgaagagaacgaagatattctcgaggacatcgtcttgacgttaaccttatttgaggatcgtgaaatgattgaggaacgcctcaaaacttatgcccacctgttcgacgataaggtgatgaagcagctgaaacgtcggcgctacacaggatggggccgcttgagtcgcaaacttattaacggaatccgtgacaagcaatccggcaaaacgattctggatttcttgaagtcggacggatttgctaatcgcaacttcatgcagttgatccatgatgactccctgacttttaaagaggatattcaaaaggcgcaggttagtggtcaaggcgacagcttacacgaacacatcgcaaatttggctggttcgccggccattaaaaaggggatcctccagaccgtgaaagttgtagatgagcttgttaaggtcatgggtcgtcataagcccgaaaacatcgtgattgaaatggcgcgggagaatcaaacgacccagaaaggacaaaagaatagccgtgaacggatgaagcggatcgaggaaggcattaaagagctggggtctcaaatcttgaaggaacaccctgtggagaacactcagctccaaaatgaaaaactttacctgtactatttgcagaacggacgcgatatgtacgtggaccaagagttggatattaatcggctgagtgactacgacgttgatcatatcgtcccgcagagcttcctcaaagacgattctattgacaataaggtactgacgcgctctgataaaaaccgtggtaagtcggacaacgtgccctccgaagaggttgtgaaaaagatgaaaaattattggcgccagcttttaaacgcgaagctgatcacacaacgtaaattcgataatttgaccaaggctgaacggggtggcctgagcgagttagataaggcaggatttattaaacgccagttagtggagactcgtcaaatcaccaaacatgtcgcgcagattttggacagccggatgaacaccaagtacgatgaaaatgacaaactgatccgtgaggtgaaagtcattactctgaagtccaaattagttagtgatttccggaaggactttcaattctacaaagtccgtgaaattaataactatcatcacgcacatgacgcgtacctgaatgcagtggttgggaccgcccttatcaagaaatatcctaagctggagtcggagtttgtctatggcgactataaggtatacgatgttcgcaaaatgattgcgaaatctgagcaggagatcggtaaggcaaccgcaaaatatttcttttactcaaacattatgaatttctttaagacagaaatcactctggccaacggggagattcgcaaacgtccgttgatcgaaacaaacggcgagactggcgaaattgtttgggacaaagggcgtgatttcgcgacggtgcgcaaggtactgagcatgcctcaagtcaatattgttaagaaaaccgaagtgcagacgggcgggttttccaaggaaagcatcttacccaaacgtaattcagataaacttattgcacgcaaaaaggactgggatccgaaaaagtatggaggcttcgacagtccaaccgtagcctactctgttctcgttgtagcgaaagtagaaaagggtaaatccaagaaactgaaatctgtcaaggagttgcttggaatcaccattatggagcgtagctccttcgagaagaacccgattgactttctggaagccaaaggatataaagaggtcaagaaagatcttatcattaagctgcctaagtattcactcttcgagctggaaaatggtcgtaaacgcatgctcgcttctgccggcgagttgcagaagggcaatgaattagcacttccatcaaagtacgttaacttcctgtatttggccagccattacgagaaactgaaggggtctccagaggacaacgaacagaaacaattatttgtagagcagcacaagcattatcttgatgaaatcattgagcaaatttccgaattcagtaaacgcgtaatcctggccgatgcaaacctcgacaaggtgctgagcgcttacaataagcatcgcgacaaacctatccgtgagcaggctgaaaatatcattcacctgttcacattaacgaacctgggcgctccggccgcttttaaatatttcgacacgacaatcgaccgtaagcgctataccagtacgaaagaagtgttggatgcgacccttattcaccagtcaattacaggattatatgagacccgtatcgaccttagccaattaggtggggatgcggccccgaagaaaaaacgcaaagtggatccgaagaaaaaacgcaaagtggcggccgcactcgagcaccaccaccaccaccactga
含有v5表位标签和2个nls序列的从经密码子优化以在人细胞中表达的dna表达的s.pycas9氨基酸序列(seqidno.:410)。
mgkpipnpllgldstapkkkrkvgihgvpaadkkysigldigtnsvgwavitdeykvpskkfkvlgntdrhsikknligallfdsgetaeatrlkrtarrrytrrknricylqeifsnemakvddsffhrleesflveedkkherhpifgnivdevayhekyptiyhlrkklvdstdkadlrliylalahmikfrghfliegdlnpdnsdvdklfiqlvqtynqlfeenpinasgvdakailsarlsksrrlenliaqlpgekknglfgnlialslgltpnfksnfdlaedaklqlskdtydddldnllaqigdqyadlflaaknlsdaillsdilrvnteitkaplsasmikrydehhqdltllkalvrqqlpekykeiffdqskngyagyidggasqeefykfikpilekmdgteellvklnredllrkqrtfdngsiphqihlgelhailrrqedfypflkdnrekiekiltfripyyvgplargnsrfawmtrkseetitpwnfeevvdkgasaqsfiermtnfdknlpnekvlpkhsllyeyftvyneltkvkyvtegmrkpaflsgeqkkaivdllfktnrkvtvkqlkedyfkkiecfdsveisgvedrfnaslgtyhdllkiikdkdfldneenediledivltltlfedremieerlktyahlfddkvmkqlkrrrytgwgrlsrklingirdkqsgktildflksdgfanrnfmqlihddsltfkediqkaqvsgqgdslhehianlagspaikkgilqtvkvvdelvkvmgrhkpeniviemarenqttqkgqknsrermkrieegikelgsqilkehpventqlqneklylyylqngrdmyvdqeldinrlsdydvdhivpqsflkddsidnkvltrsdknrgksdnvpseevvkkmknywrqllnaklitqrkfdnltkaergglseldkagfikrqlvetrqitkhvaqildsrmntkydendklirevkvitlksklvsdfrkdfqfykvreinnyhhahdaylnavvgtalikkypklesefvygdykvydvrkmiakseqeigkatakyffysnimnffkteitlangeirkrplietngetgeivwdkgrdfatvrkvlsmpqvnivkktevqtggfskesilpkrnsdkliarkkdwdpkkyggfdsptvaysvlvvakvekgkskklksvkellgitimerssfeknpidfleakgykevkkdliiklpkyslfelengrkrmlasagelqkgnelalpskyvnflylashyeklkgspedneqkqlfveqhkhyldeiieqisefskrviladanldkvlsaynkhrdkpireqaeniihlftltnlgapaafkyfdttidrkrytstkevldatlihqsitglyetridlsqlggdsradpkkkrkvefhhtglvdpssvpslslnr
含有v5表位标签和2个nls序列的经密码子优化以在人细胞中表达的s.pycas9dna序列(seqidno.:411)。
atgggcaagcccatccctaaccccctgttggggctggacagcaccgctcccaaaaagaaaaggaaggtgggcattcacggcgtgcctgcggccgacaaaaagtacagcatcggccttgatatcggcaccaatagcgtgggctgggccgttatcacagacgaatacaaggtacccagcaagaagttcaaggtgctggggaatacagacaggcactctatcaagaaaaaccttatcggggctctgctgtttgactcaggcgagaccgccgaggccaccaggttgaagaggaccgcaaggcgaaggtacacccggaggaagaacaggatctgctatctgcaggagatcttcagcaacgagatggccaaggtggacgacagcttcttccacaggctggaggagagcttccttgtcgaggaggataagaagcacgaacgacaccccatcttcggcaacatagtcgacgaggtcgcttatcacgagaagtaccccaccatctaccacctgcgaaagaaattggtggatagcaccgataaagccgacttgcgacttatctacttggctctggcgcacatgattaagttcaggggccacttcctgatcgagggcgaccttaaccccgacaacagtgacgtagacaaattgttcatccagcttgtacagacctataaccagctgttcgaggaaaaccctattaacgccagcggggtggatgcgaaggccatacttagcgccaggctgagcaaaagcaggcgcttggagaacctgatagcccagctgcccggtgaaaagaagaacggcctcttcggtaatctgattgccctgagcctgggcctgacccccaacttcaagagcaacttcgacctggcagaagatgccaagctgcagttgagtaaggacacctatgacgacgacttggacaatctgctcgcccaaatcggcgaccagtacgctgacctgttcctcgccgccaagaacctttctgacgcaatcctgcttagcgatatccttagggtgaacacagagatcaccaaggcccccctgagcgccagcatgatcaagaggtacgacgagcaccatcaggacctgacccttctgaaggccctggtgaggcagcaactgcccgagaagtacaaggagatctttttcgaccagagcaagaacggctacgccggctacatcgacggcggagccagccaagaggagttctacaagttcatcaagcccatcctggagaagatggatggcaccgaggagctgctggtgaagctgaacagggaagatttgctccggaagcagaggacctttgacaacggtagcatcccccaccagatccacctgggcgagctgcacgcaatactgaggcgacaggaggatttctaccccttcctcaaggacaatagggagaaaatcgaaaagattctgaccttcaggatcccctactacgtgggccctcttgccaggggcaacagccgattcgcttggatgacaagaaagagcgaggagaccatcaccccctggaacttcgaggaagtggtggacaaaggagcaagcgcgcagtctttcatcgaacggatgaccaatttcgacaaaaacctgcctaacgagaaggtgctgcccaagcacagcctgctttacgagtacttcaccgtgtacaacgagctcaccaaggtgaaatatgtgaccgagggcatgcgaaaacccgctttcctgagcggcgagcagaagaaggccatcgtggacctgctgttcaagaccaacaggaaggtgaccgtgaagcagctgaaggaggactacttcaagaagatcgagtgctttgatagcgtggaaataagcggcgtggaggacaggttcaacgccagcctgggcacctaccacgacttgttgaagataatcaaagacaaggatttcctggataatgaggagaacgaggatatactcgaggacatcgtgctgactttgaccctgtttgaggaccgagagatgattgaagaaaggctcaaaacctacgcccacctgttcgacgacaaagtgatgaaacaactgaagagacgaagatacaccggctggggcagactgtccaggaagctcatcaacggcattagggacaagcagagcggcaagaccatcctggatttcctgaagtccgacggcttcgccaaccgaaacttcatgcagctgattcacgatgacagcttgaccttcaaggaggacatccagaaggcccaggttagcggccagggcgactccctgcacgaacatattgcaaacctggcaggctcccctgcgatcaagaagggcatactgcagaccgttaaggttgtggacgaattggtcaaggtcatgggcaggcacaagcccgaaaacatagttatagagatggccagagagaaccagaccacccaaaagggccagaagaacagccgggagcgcatgaaaaggatcgaggagggtatcaaggaactcggaagccagatcctcaaagagcaccccgtggagaatacccagctccagaacgagaagctgtacctgtactacctgcagaacggcagggacatgtacgttgaccaggagttggacatcaacaggctttcagactatgacgtggatcacatagtgccccagagctttcttaaagacgatagcatcgacaacaaggtcctgacccgctccgacaaaaacaggggcaaaagcgacaacgtgccaagcgaagaggtggttaaaaagatgaagaactactggaggcaactgctcaacgcgaaattgatcacccagagaaagttcgataacctgaccaaggccgagaggggcggactctccgaacttgacaaagcgggcttcataaagaggcagctggtcgagacccgacagatcacgaagcacgtggcccaaatcctcgacagcagaatgaataccaagtacgatgagaatgacaaactcatcagggaagtgaaagtgattaccctgaagagcaagttggtgtccgactttcgcaaagatttccagttctacaaggtgagggagatcaacaactaccaccatgcccacgacgcatacctgaacgccgtggtcggcaccgccctgattaagaagtatccaaagctggagtccgaatttgtctacggcgactacaaagtttacgatgtgaggaagatgatcgctaagagcgaacaggagatcggcaaggccaccgctaagtatttcttctacagcaacatcatgaactttttcaagaccgagatcacacttgccaacggcgaaatcaggaagaggccgcttatcgagaccaacggtgagaccggcgagatcgtgtgggacaagggcagggacttcgccaccgtgaggaaagtcctgagcatgccccaggtgaatattgtgaaaaaaactgaggtgcagacaggcggctttagcaaggaatccatcctgcccaagaggaacagcgacaagctgatcgcccggaagaaggactgggaccctaagaagtatggaggcttcgacagccccaccgtagcctacagcgtgctggtggtcgcgaaggtagagaaggggaagagcaagaaactgaagagcgtgaaggagctgctcggcataaccatcatggagaggtccagctttgagaagaaccccattgactttttggaagccaagggctacaaagaggtcaaaaaggacctgatcatcaaactccccaagtactccctgtttgaattggagaacggcagaaagaggatgctggcgagcgctggggaactgcaaaagggcaacgaactggcgctgcccagcaagtacgtgaattttctgtacctggcgtcccactacgaaaagctgaaaggcagccccgaggacaacgagcagaagcagctgttcgtggagcagcacaagcattacctggacgagataatcgagcaaatcagcgagttcagcaagagggtgattctggccgacgcgaacctggataaggtcctcagcgcctacaacaagcaccgagacaaacccatcagggagcaggccgagaatatcatacacctgttcaccctgacaaatctgggcgcacctgcggcattcaaatacttcgataccaccatcgacaggaaaaggtacactagcactaaggaggtgctggatgccaccttgatccaccagtccattaccggcctgtatgagaccaggatcgacctgagccagcttggaggcgactctagggcggacccaaaaaagaaaaggaaggtggaattccaccacactggactagtggatccgagctcggtaccaagcttaagtttaaaccgctga
经密码子优化以在人细胞中作为t7rna聚合酶表达盒表达的s.pycas9dna序列(seqidno.:412)。该序列含有t7rna聚合酶启动子、v5表位标签、核定位信号、密码子优化的cas9序列、第二核定位信号,和具有多聚腺苷酸化信号的bgh(牛生长激素)基因3'-utr元件。
taatacgactcactatagggagacccaagctggctagcgtttaaacgggccctctagactcgagcggccgccaccatgggcaagcccatccctaaccccctgttggggctggacagcaccgctcccaaaaagaaaaggaaggtgggcattcacggcgtgcctgcggccgacaaaaagtacagcatcggccttgatatcggcaccaatagcgtgggctgggccgttatcacagacgaatacaaggtacccagcaagaagttcaaggtgctggggaatacagacaggcactctatcaagaaaaaccttatcggggctctgctgtttgactcaggcgagaccgccgaggccaccaggttgaagaggaccgcaaggcgaaggtacacccggaggaagaacaggatctgctatctgcaggagatcttcagcaacgagatggccaaggtggacgacagcttcttccacaggctggaggagagcttccttgtcgaggaggataagaagcacgaacgacaccccatcttcggcaacatagtcgacgaggtcgcttatcacgagaagtaccccaccatctaccacctgcgaaagaaattggtggatagcaccgataaagccgacttgcgacttatctacttggctctggcgcacatgattaagttcaggggccacttcctgatcgagggcgaccttaaccccgacaacagtgacgtagacaaattgttcatccagcttgtacagacctataaccagctgttcgaggaaaaccctattaacgccagcggggtggatgcgaaggccatacttagcgccaggctgagcaaaagcaggcgcttggagaacctgatagcccagctgcccggtgaaaagaagaacggcctcttcggtaatctgattgccctgagcctgggcctgacccccaacttcaagagcaacttcgacctggcagaagatgccaagctgcagttgagtaaggacacctatgacgacgacttggacaatctgctcgcccaaatcggcgaccagtacgctgacctgttcctcgccgccaagaacctttctgacgcaatcctgcttagcgatatccttagggtgaacacagagatcaccaaggcccccctgagcgccagcatgatcaagaggtacgacgagcaccatcaggacctgacccttctgaaggccctggtgaggcagcaactgcccgagaagtacaaggagatctttttcgaccagagcaagaacggctacgccggctacatcgacggcggagccagccaagaggagttctacaagttcatcaagcccatcctggagaagatggatggcaccgaggagctgctggtgaagctgaacagggaagatttgctccggaagcagaggacctttgacaacggtagcatcccccaccagatccacctgggcgagctgcacgcaatactgaggcgacaggaggatttctaccccttcctcaaggacaatagggagaaaatcgaaaagattctgaccttcaggatcccctactacgtgggccctcttgccaggggcaacagccgattcgcttggatgacaagaaagagcgaggagaccatcaccccctggaacttcgaggaagtggtggacaaaggagcaagcgcgcagtctttcatcgaacggatgaccaatttcgacaaaaacctgcctaacgagaaggtgctgcccaagcacagcctgctttacgagtacttcaccgtgtacaacgagctcaccaaggtgaaatatgtgaccgagggcatgcgaaaacccgctttcctgagcggcgagcagaagaaggccatcgtggacctgctgttcaagaccaacaggaaggtgaccgtgaagcagctgaaggaggactacttcaagaagatcgagtgctttgatagcgtggaaataagcggcgtggaggacaggttcaacgccagcctgggcacctaccacgacttgttgaagataatcaaagacaaggatttcctggataatgaggagaacgaggatatactcgaggacatcgtgctgactttgaccctgtttgaggaccgagagatgattgaagaaaggctcaaaacctacgcccacctgttcgacgacaaagtgatgaaacaactgaagagacgaagatacaccggctggggcagactgtccaggaagctcatcaacggcattagggacaagcagagcggcaagaccatcctggatttcctgaagtccgacggcttcgccaaccgaaacttcatgcagctgattcacgatgacagcttgaccttcaaggaggacatccagaaggcccaggttagcggccagggcgactccctgcacgaacatattgcaaacctggcaggctcccctgcgatcaagaagggcatactgcagaccgttaaggttgtggacgaattggtcaaggtcatgggcaggcacaagcccgaaaacatagttatagagatggccagagagaaccagaccacccaaaagggccagaagaacagccgggagcgcatgaaaaggatcgaggagggtatcaaggaactcggaagccagatcctcaaagagcaccccgtggagaatacccagctccagaacgagaagctgtacctgtactacctgcagaacggcagggacatgtacgttgaccaggagttggacatcaacaggctttcagactatgacgtggatcacatagtgccccagagctttcttaaagacgatagcatcgacaacaaggtcctgacccgctccgacaaaaacaggggcaaaagcgacaacgtgccaagcgaagaggtggttaaaaagatgaagaactactggaggcaactgctcaacgcgaaattgatcacccagagaaagttcgataacctgaccaaggccgagaggggcggactctccgaacttgacaaagcgggcttcataaagaggcagctggtcgagacccgacagatcacgaagcacgtggcccaaatcctcgacagcagaatgaataccaagtacgatgagaatgacaaactcatcagggaagtgaaagtgattaccctgaagagcaagttggtgtccgactttcgcaaagatttccagttctacaaggtgagggagatcaacaactaccaccatgcccacgacgcatacctgaacgccgtggtcggcaccgccctgattaagaagtatccaaagctggagtccgaatttgtctacggcgactacaaagtttacgatgtgaggaagatgatcgctaagagcgaacaggagatcggcaaggccaccgctaagtatttcttctacagcaacatcatgaactttttcaagaccgagatcacacttgccaacggcgaaatcaggaagaggccgcttatcgagaccaacggtgagaccggcgagatcgtgtgggacaagggcagggacttcgccaccgtgaggaaagtcctgagcatgccccaggtgaatattgtgaaaaaaactgaggtgcagacaggcggctttagcaaggaatccatcctgcccaagaggaacagcgacaagctgatcgcccggaagaaggactgggaccctaagaagtatggaggcttcgacagccccaccgtagcctacagcgtgctggtggtcgcgaaggtagagaaggggaagagcaagaaactgaagagcgtgaaggagctgctcggcataaccatcatggagaggtccagctttgagaagaaccccattgactttttggaagccaagggctacaaagaggtcaaaaaggacctgatcatcaaactccccaagtactccctgtttgaattggagaacggcagaaagaggatgctggcgagcgctggggaactgcaaaagggcaacgaactggcgctgcccagcaagtacgtgaattttctgtacctggcgtcccactacgaaaagctgaaaggcagccccgaggacaacgagcagaagcagctgttcgtggagcagcacaagcattacctggacgagataatcgagcaaatcagcgagttcagcaagagggtgattctggccgacgcgaacctggataaggtcctcagcgcctacaacaagcaccgagacaaacccatcagggagcaggccgagaatatcatacacctgttcaccctgacaaatctgggcgcacctgcggcattcaaatacttcgataccaccatcgacaggaaaaggtacactagcactaaggaggtgctggatgccaccttgatccaccagtccattaccggcctgtatgagaccaggatcgacctgagccagcttggaggcgactctagggcggacccaaaaaagaaaaggaaggtggaattccaccacactggactagtggatccgagctcggtaccaagcttaagtttaaaccgctgatcagcctcgactgtgccttctagttgccagccatctgttgtttgcccctcccccgtgccttccttgaccctggaaggtgccactcccactgtcctttcctaataaaatgaggaaattgcatcgcattgtctgagtaggtgtcattctattctggggggtggggtggggcaggacagcaagggggaggattgggaagacaatagcaggcatgctggggatgcggtgggctctatggc
从表达盒(seqidno.:412)制备的s.pycas9mrna(seqidno.:413)。该序列含有v5表位标签、核定位信号、密码子优化的cas9序列、第二核定位信号,以及bgh(牛生长激素)基因3'-utr元件和聚a尾。
gggagacccaagcuggcuagcguuuaaacgggcccucuagacucgagcggccgccaccaugggcaagcccaucccuaacccccuguuggggcuggacagcaccgcucccaaaaagaaaaggaaggugggcauucacggcgugccugcggccgacaaaaaguacagcaucggccuugauaucggcaccaauagcgugggcugggccguuaucacagacgaauacaagguacccagcaagaaguucaaggugcuggggaauacagacaggcacucuaucaagaaaaaccuuaucggggcucugcuguuugacucaggcgagaccgccgaggccaccagguugaagaggaccgcaaggcgaagguacacccggaggaagaacaggaucugcuaucugcaggagaucuucagcaacgagauggccaagguggacgacagcuucuuccacaggcuggaggagagcuuccuugucgaggaggauaagaagcacgaacgacaccccaucuucggcaacauagucgacgaggucgcuuaucacgagaaguaccccaccaucuaccaccugcgaaagaaauugguggauagcaccgauaaagccgacuugcgacuuaucuacuuggcucuggcgcacaugauuaaguucaggggccacuuccugaucgagggcgaccuuaaccccgacaacagugacguagacaaauuguucauccagcuuguacagaccuauaaccagcuguucgaggaaaacccuauuaacgccagcgggguggaugcgaaggccauacuuagcgccaggcugagcaaaagcaggcgcuuggagaaccugauagcccagcugcccggugaaaagaagaacggccucuucgguaaucugauugcccugagccugggccugacccccaacuucaagagcaacuucgaccuggcagaagaugccaagcugcaguugaguaaggacaccuaugacgacgacuuggacaaucugcucgcccaaaucggcgaccaguacgcugaccuguuccucgccgccaagaaccuuucugacgcaauccugcuuagcgauauccuuagggugaacacagagaucaccaaggccccccugagcgccagcaugaucaagagguacgacgagcaccaucaggaccugacccuucugaaggcccuggugaggcagcaacugcccgagaaguacaaggagaucuuuuucgaccagagcaagaacggcuacgccggcuacaucgacggcggagccagccaagaggaguucuacaaguucaucaagcccauccuggagaagauggauggcaccgaggagcugcuggugaagcugaacagggaagauuugcuccggaagcagaggaccuuugacaacgguagcaucccccaccagauccaccugggcgagcugcacgcaauacugaggcgacaggaggauuucuaccccuuccucaaggacaauagggagaaaaucgaaaagauucugaccuucaggauccccuacuacgugggcccucuugccaggggcaacagccgauucgcuuggaugacaagaaagagcgaggagaccaucacccccuggaacuucgaggaagugguggacaaaggagcaagcgcgcagucuuucaucgaacggaugaccaauuucgacaaaaaccugccuaacgagaaggugcugcccaagcacagccugcuuuacgaguacuucaccguguacaacgagcucaccaaggugaaauaugugaccgagggcaugcgaaaacccgcuuuccugagcggcgagcagaagaaggccaucguggaccugcuguucaagaccaacaggaaggugaccgugaagcagcugaaggaggacuacuucaagaagaucgagugcuuugauagcguggaaauaagcggcguggaggacagguucaacgccagccugggcaccuaccacgacuuguugaagauaaucaaagacaaggauuuccuggauaaugaggagaacgaggauauacucgaggacaucgugcugacuuugacccuguuugaggaccgagagaugauugaagaaaggcucaaaaccuacgcccaccuguucgacgacaaagugaugaaacaacugaagagacgaagauacaccggcuggggcagacuguccaggaagcucaucaacggcauuagggacaagcagagcggcaagaccauccuggauuuccugaaguccgacggcuucgccaaccgaaacuucaugcagcugauucacgaugacagcuugaccuucaaggaggacauccagaaggcccagguuagcggccagggcgacucccugcacgaacauauugcaaaccuggcaggcuccccugcgaucaagaagggcauacugcagaccguuaagguuguggacgaauuggucaaggucaugggcaggcacaagcccgaaaacauaguuauagagauggccagagagaaccagaccacccaaaagggccagaagaacagccgggagcgcaugaaaaggaucgaggaggguaucaaggaacucggaagccagauccucaaagagcaccccguggagaauacccagcuccagaacgagaagcuguaccuguacuaccugcagaacggcagggacauguacguugaccaggaguuggacaucaacaggcuuucagacuaugacguggaucacauagugccccagagcuuucuuaaagacgauagcaucgacaacaagguccugacccgcuccgacaaaaacaggggcaaaagcgacaacgugccaagcgaagaggugguuaaaaagaugaagaacuacuggaggcaacugcucaacgcgaaauugaucacccagagaaaguucgauaaccugaccaaggccgagaggggcggacucuccgaacuugacaaagcgggcuucauaaagaggcagcuggucgagacccgacagaucacgaagcacguggcccaaauccucgacagcagaaugaauaccaaguacgaugagaaugacaaacucaucagggaagugaaagugauuacccugaagagcaaguugguguccgacuuucgcaaagauuuccaguucuacaaggugagggagaucaacaacuaccaccaugcccacgacgcauaccugaacgccguggucggcaccgcccugauuaagaaguauccaaagcuggaguccgaauuugucuacggcgacuacaaaguuuacgaugugaggaagaugaucgcuaagagcgaacaggagaucggcaaggccaccgcuaaguauuucuucuacagcaacaucaugaacuuuuucaagaccgagaucacacuugccaacggcgaaaucaggaagaggccgcuuaucgagaccaacggugagaccggcgagaucgugugggacaagggcagggacuucgccaccgugaggaaaguccugagcaugccccaggugaauauugugaaaaaaacugaggugcagacaggcggcuuuagcaaggaauccauccugcccaagaggaacagcgacaagcugaucgcccggaagaaggacugggacccuaagaaguauggaggcuucgacagccccaccguagccuacagcgugcugguggucgcgaagguagagaaggggaagagcaagaaacugaagagcgugaaggagcugcucggcauaaccaucauggagagguccagcuuugagaagaaccccauugacuuuuuggaagccaagggcuacaaagaggucaaaaaggaccugaucaucaaacuccccaaguacucccuguuugaauuggagaacggcagaaagaggaugcuggcgagcgcuggggaacugcaaaagggcaacgaacuggcgcugcccagcaaguacgugaauuuucuguaccuggcgucccacuacgaaaagcugaaaggcagccccgaggacaacgagcagaagcagcuguucguggagcagcacaagcauuaccuggacgagauaaucgagcaaaucagcgaguucagcaagagggugauucuggccgacgcgaaccuggauaagguccucagcgccuacaacaagcaccgagacaaacccaucagggagcaggccgagaauaucauacaccuguucacccugacaaaucugggcgcaccugcggcauucaaauacuucgauaccaccaucgacaggaaaagguacacuagcacuaaggaggugcuggaugccaccuugauccaccaguccauuaccggccuguaugagaccaggaucgaccugagccagcuuggaggcgacucuagggcggacccaaaaaagaaaaggaagguggaauuccaccacacuggacuaguggauccgagcucgguaccaagcuuaaguuuaaaccgcugaucagccucgacugugccuucuaguugccagccaucuguuguuugccccucccccgugccuuccuugacccuggaaggugccacucccacuguccuuuccuaauaaaaugaggaaauugcaucgcauugucugaguaggugucauucuauucuggggggugggguggggcaggacagcaagggggaggauugggaagacaauagcaggcaugcuggggaugcggugggcucuauggc–polya
实施例12
以下实施例表明,当与未修饰的ivtsgrna相比时,本发明的截短的化学修饰的crrna:tracrrna复合物减少了哺乳动物细胞中的先天免疫系统的刺激。
哺乳动物细胞具有预期鉴定和响应外来rna作为抗病毒免疫的一部分的多种受体。这包括诸如tlr-3、tlr-7、tlr8、rig-i、mda5、oas、pkr等受体。广义而言,哺乳动物细胞中存在的短的或含有化学修饰的rna(例如2'omerna)逃避检测或者不如长的未修饰的rna具刺激性。本实施例将当用本发明的截短的未修饰的或截短的修饰的crrna:tracrrna复合物转染哺乳动物hek293细胞时2个免疫反应相关基因(ifit1和ifitm1)的刺激水平与商业ivtsgrna(thermofisherscientific,waltham,ma)进行比较。
使用对人hprt1位点38285具特异性的crispr向导rna。序列示于下表11中。如上文实施例2中所概述,将未修饰的crrna:tracrrna复合物(seqidno.:48和2)、修饰的crrna:tracrrna复合物(seqidno.:178和100)和sgrna(seqidno.:414)以30nm浓度转染到hek-cas9细胞中。使用sv96总rna分离试剂盒(promega,madison,wi)在转染24小时后制备rna。使用随机六聚体和寡dt引发,根据制造商的说明书使用150ng总rna与superscripttm-ii逆转录酶(invitrogen,carlsbad,ca)来合成cdna。转染实验都进行最少三次。
对于每10μl反应物使用10ngcdna与immolasetmdna聚合酶(bioline,randolph,ma)、200nm引物和200nm探针来进行定量实时pcr。使用的循环条件是:95℃持续10分钟,接着是40个循环的2步pcr,其中95℃持续15秒并且60℃持续1分钟。使用abiprismtm7900序列检测器(appliedbiosystemsinc.,fostercity,ca)进行pcr和荧光测量。所有反应都使用双色复用一式三份地进行。表达数据相对于两个内部对照基因的平均值进行归一化。拷贝数标准是所有测定的线性化克隆扩增子。相对于标准来外推未知数,以建立绝对的定量测量。家用内部对照归一化测定是hprt1(引物和探针seqidno.:415-417)和sfrs9(引物和探针seqidno.:418-420)。免疫激活途径测定是ifitm1(引物和探针seqidno.:421-423)和ifit1(引物和探针seqidno.:424-426)。使用非转染细胞作为基线将结果归一化并示于图13中。
表11.在实施例12中的免疫激活实验中使用的核酸试剂。
1化合物i是具有式seqidno:417-(zen)-seqidno:441的寡核苷酸。
2化合物ii是具有式seqidno:420-(zen)-seqidno:442的寡核苷酸。
3化合物iii是具有式seqidno:423-(zen)-seqidno:443的寡核苷酸。
4化合物iv是具有式seqidno:426-(zen)-seqidno:444的寡核苷酸。
寡核苷酸序列以5'-3'示出。大写字母=dna;小写字母=rna;加下划线=2'-o-甲基rna;*=硫代磷酸酯核苷酸间的键;ppp=三磷酸酯;zen=萘基-偶氮修饰剂,暗淬灭剂;fam=6-羧基荧光素;hex=六氯荧光素。
用未修饰或化学修饰的截短的crrna:tracrrna复合物进行处理不会导致ifit1或ifitm1表达超过基线的可检测的增加。相比之下,用较长的ivtsgrna进行处理导致ifitm1的45倍诱导和ifit1的220倍诱导。因此,使用本发明的短crrna:tracrrna复合物不存在使用sgrna发生的先天免疫系统的显著刺激。
实施例13
以下实施例将实施例6和7中鉴定的修饰模式组合,其特别有效地展示了在哺乳动物crispr基因组编辑应用中以高效率表现的新的高度修饰的crrna和tracrrna组合物。
合成了一系列crrna和tracrrna(表12),其具有如所示的化学修饰。crrna采用位于5’末端的靶向人hprt1基因中的相同位点(38285)的20个碱基前间区序列结构域与位于3’末端的16个碱基tracrrna结合结构域。使用tracrrna序列的67个核苷酸或62个核苷酸截短形式,合成了具有所示化学修饰的tracrrna。将表12中列出的crrna和tracrrna如所示进行配对,并且以30nm浓度转染到hek-cas9细胞中并如先前实施例中所述进行处理。通过比较使用t7ei错配核酸内切酶切割得到的hprt1基因中的切割率与使用片段分析仪进行的产物的定量测量来评估相对基因编辑活性。
表12:高度修饰的crrna:tracrrna复合物在哺乳动物细胞中引导cas9介导的基因编辑的活性。
寡核苷酸序列以5'-3'示出。小写字母=rna;加下划线=2'-o-甲基rna;小写字母斜体字=2'frna;*=硫代磷酸酯核苷酸间的键。对于所研究的每个剂量,每个复合物的相对功能活性由t7ei异源双链体测定中的切割%指示。
crrna:tracrrna对#1和#2显示,高度2'frna修饰的crrna(seqidno.:448,其中22/36残基被修饰,或61%)当与未修饰的tracrrna(seqidno.:2)或高度2'ome修饰的tracrrna(seqidno.:100)配对时具有高功能性。crrna:tracrrna对#3和#4显示,具有中度(seqidno.:450,其中19/67残基被修饰,或28%)或高度(seqidno.:449,其中46/67残基被修饰,或69%)2'frna修饰水平的tracrrna组合物具有高功能性。源自实施例6的信息(特别是2'ome‘步行’,seqidno.:144-162)用于鉴定可在tracrrna的内部结构域内修饰的特定残基(参见图6)。crrna:tracrrna对#5表明,非常高度修饰的tracrrna,其在这种情况下是截短的62个核苷酸设计(seqidno.:451,51/62残基被2'omerna修饰,或82%),在哺乳动物细胞中触发crispr基因组编辑时具有高效能。因此,原始89rna核苷酸野生型tracrrna已在本文优化为剩余少至11个rna残基(11/62)的形式,从而显著降低了哺乳动物先天免疫系统的基于rna的激活风险,并且将tracrrna的核酸酶敏感rna含量降至最低水平。
本文引用的所有参考文献(包括出版物、专利申请和专利)特此通过引用并入,其程度就如同每个参考文献被单独地和明确地指示是通过引用并入并且在此全文阐述一样。
除非本文另有说明或者上下文明显相矛盾,否则在描述本发明的上下文中(特别是在所附权利要求的上下文中)使用术语“一”和“所述”以及类似的指示物应被解释为涵盖单数和复数。除非另有说明,否则术语“包含”、“具有”、“包括”和“含有”将被解释为开放式术语(即,是指“包括但不限于”)。除非本文另有说明,否则本文中数值范围的描述仅仅意图用作单独提及落在该范围内的每个单独数值的简写方法,并且将每个单独的数值并入本说明书中,就如同其在本文中单独叙述一样。除非本文另有说明或者上下文明显相矛盾,否则本文所述的所有方法可以任何合适的顺序进行。除非另有说明,否则本文中提供的任何和所有的实例或示例性语言(例如,“诸如”)的使用仅旨在更好地说明本发明,而不对本发明的范围构成限制。本说明书中的语言不应被解释为指示任何未要求保护的要素对于本发明的实践是必需的。
本文描述了本发明的优选实施方案,包括本发明人已知的用于实施本发明的最佳模式。在阅读前面的描述之后,这些优选实施方案的变化对于本领域普通技术人员来说可能变得显而易见。本发明人期望本领域技术人员适当地使用这种变化,并且本发明人旨在以不同于本文具体描述的方式实施本发明。因此,本发明包括根据适用法律允许的所附权利要求中所述的主题的所有修改和等同物。此外,除非本文另有说明或者上下文明显相矛盾,否则本发明涵盖了上述要素呈其所有可能变化形式的任何组合。
序列表
<110>m·a·科林伍德
a·m·雅各比
g·r·雷蒂希
m·s·舒伯特
m·a·贝尔克
<120>基于crispr的组合物和使用方法
<130>idt01-008-us
<140>
<141>
<150>62/239,546
<151>2015-10-09
<150>62/093,588
<151>2014-12-18
<160>451
<170>patentinversion3.5
<210>1
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>1
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>2
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>2
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>3
<211>89
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>3
guuggaaccauucaaaacagcauagcaaguuaaaauaaggcuaguccguuaucaacuuga60
aaaaguggcaccgagucggugcuuuuuuu89
<210>4
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>4
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>5
<211>41
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>5
uuauauccaacacuucgugguuuuagagcuaugcuguuuug41
<210>6
<211>89
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>6
guuggaaccauucaaaacagcauagcaaguuaaaauaaggcuaguccguuaucaacuuga60
aaaaguggcaccgagucggugcuuuuuuu89
<210>7
<211>41
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>7
uuauauccaacacuucgugguuuuagagcuaugcuguuuug41
<210>8
<211>41
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>8
uuauauccaacacuucgugguuuuagagcuaugcuguuuug41
<210>9
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>9
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>10
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>10
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>11
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>11
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>12
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>12
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>13
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>13
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>14
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>14
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>15
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>15
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>16
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>16
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>17
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>17
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>18
<211>89
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>18
guuggaaccauucaaaacagcauagcaaguuaaaauaaggcuaguccguuaucaacuuga60
aaaaguggcaccgagucggugcuuuuuuu89
<210>19
<211>89
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>19
guuggaaccauucaaaacagcauagcaaguuaaaauaaggcuaguccguuaucaacuuga60
aaaaguggcaccgagucggugcuuuuuuu89
<210>20
<211>89
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>20
guuggaaccauucaaaacagcauagcaaguuaaaauaaggcuaguccguuaucaacuuga60
aaaaguggcaccgagucggugcuuuuuuu89
<210>21
<211>89
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>21
guuggaaccauucaaaacagcauagcaaguuaaaauaaggcuaguccguuaucaacuuga60
aaaaguggcaccgagucggugcuuuuuuu89
<210>22
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>22
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>23
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>23
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>24
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>24
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>25
<211>41
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>25
uuauauccaacacuucgugguuuuagagcuaugcuguuuug41
<210>26
<211>41
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>26
uuauauccaacacuucgugguuuuagagcuaugcuguuuug41
<210>27
<211>938
<212>dna
<213>homosapiens
<400>27
gaatgttgtgataaaaggtgatgctcacctctcccacacccttttatagtttagggattg60
tatttccaaggtttctagactgagagcccttttcatctttgctcattgacactctgtacc120
cattaatcctccttattagctccccttcaatggacacatgggtagtcagggtgcaggtct180
cagaactgtccttcaggttccaggtgatcaaccaagtgccttgtctgtagtgtcaactca240
ttgctgccccttcctagtaatccccataatttagctctccatttcatagtctttccttgg300
gtgtgttaaaagtgaccatggtacactcagcacggatgaaatgaaacagtgtttagaaac360
gtcagtcttctcttttgtaatgccctgtagtctctctgtatgttatatgtcacattttgt420
aattaacagcttgctggtgaaaaggaccccacgaagtgttggatataagccagactgtaa480
gtgaattactttttttgtcaatcatttaaccatctttaacctaaaagagttttatgtgaa540
atggcttataattgcttagagaatatttgtagagaggcacatttgccagtattagattta600
aaagtgatgttttctttatctaaatgatgaattatgattctttttagttgttggatttga660
aattccagacaagtttgttgtaggatatgcccttgactataatgaatacttcagggattt720
gaatgtaagtaattgcttctttttctcactcatttttcaaaacacgcataaaaatttagg780
aaagagaattgttttctccttccagcacctcataatttgaacagactgatggttcccatt840
agtcacataaagctgtagtctagtacagacgtccttagaactggaacctggccaggctag900
ggtgacacttcttgttggctgaaatagttgaacagctt938
<210>28
<211>27
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
primer
<400>28
aagaatgttgtgataaaaggtgatgct27
<210>29
<211>24
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
primer
<400>29
acacatccatgggacttctgcctc24
<210>30
<211>74
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>30
caaaacagcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcacc60
gagucggugcuuuu74
<210>31
<211>70
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>31
aacagcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgag60
ucggugcuuu70
<210>32
<211>65
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>32
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcu65
<210>33
<211>63
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>33
cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucggu60
gcu63
<210>34
<211>55
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>34
agcauagcaaguuaaaauaguuaucaacuugaaaaaguggcaccgagucggugcu55
<210>35
<211>49
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>35
agcauagcaaguuaaaauaaacuugaaaaaguggcaccgagucggugcu49
<210>36
<211>64
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>36
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugc64
<210>37
<211>63
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>37
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gug63
<210>38
<211>62
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>38
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gu62
<210>39
<211>61
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>39
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
g61
<210>40
<211>58
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>40
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagugccgagucgg58
<210>41
<211>59
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>41
agcauagcaaguuaaaauaaggcuaguccaacuugaaaaaguggcaccgagucggugcu59
<210>42
<211>55
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>42
agcauagcaaguuaaaauaaggcuaguccaacuugaaaaaguggcaccgagucgg55
<210>43
<211>52
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>43
agcauagcaaguuaaaauaaggcuaguccaacuugaaaaagugccgagucgg52
<210>44
<211>49
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>44
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaagug49
<210>45
<211>52
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>45
agcauagcaaguuaaaauaaggcuaguccguuaucagcaccgagucggugcu52
<210>46
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>46
cuuauauccaacacuucgugguuuuagagcuaugcuguuuug42
<210>47
<211>39
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>47
cuuauauccaacacuucgugguuuuagagcuaugcuguu39
<210>48
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>48
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>49
<211>34
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>49
cuuauauccaacacuucgugguuuuagagcuaug34
<210>50
<211>74
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>50
caaaacagcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcacc60
gagucggugcuuuu74
<210>51
<211>70
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>51
aacagcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgag60
ucggugcuuu70
<210>52
<211>65
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>52
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcu65
<210>53
<211>63
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>53
cauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucggu60
gcu63
<210>54
<211>34
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>54
uauauccaacacuucgugguuuuagagcuaugcu34
<210>55
<211>33
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>55
auauccaacacuucgugguuuuagagcuaugcu33
<210>56
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>56
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>57
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>57
auuauggggauuacuaggaguuuuagagcuaugcu35
<210>58
<211>34
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>58
uuauggggauuacuaggaguuuuagagcuaugcu34
<210>59
<211>33
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>59
uauggggauuacuaggaguuuuagagcuaugcu33
<210>60
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>60
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>61
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>61
uuucacauaaaacucuuuuguuuuagagcuaugcu35
<210>62
<211>34
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>62
uucacauaaaacucuuuuguuuuagagcuaugcu34
<210>63
<211>33
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>63
ucacauaaaacucuuuuguuuuagagcuaugcu33
<210>64
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>64
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>65
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>65
ccauuucauagucuuuccuguuuuagagcuaugcu35
<210>66
<211>34
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>66
cauuucauagucuuuccuguuuuagagcuaugcu34
<210>67
<211>33
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>67
auuucauagucuuuccuguuuuagagcuaugcu33
<210>68
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>68
uccauuucauagucuuuccuguuuuagagcuaugcuguuuug42
<210>69
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>69
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>70
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>70
uuuuguaauuaacagcuugcguuuuagagcuaugcuguuuug42
<210>71
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>71
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>72
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>72
cuuagagaauauuuguagagguuuuagagcuaugcuguuuug42
<210>73
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>73
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>74
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>74
uugacuauaaugaauacuucguuuuagagcuaugcuguuuug42
<210>75
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>75
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>76
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>76
caaaacacgcauaaaaauuuguuuuagagcuaugcuguuuug42
<210>77
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>77
aauuauggggauuacuaggaguuuuagagcuaugcuguuuug42
<210>78
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>78
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>79
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>79
ggucacuuuuaacacacccaguuuuagagcuaugcuguuuug42
<210>80
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>80
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>81
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>81
ggcuuauauccaacacuucgguuuuagagcuaugcuguuuug42
<210>82
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>82
auuucacauaaaacucuuuuguuuuagagcuaugcuguuuug42
<210>83
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>83
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>84
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>84
ucaaauuaugaggugcuggaguuuuagagcuaugcuguuuug42
<210>85
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>85
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>86
<211>42
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>86
uacagcuuuaugugacuaauguuuuagagcuaugcuguuuug42
<210>87
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>87
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>88
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>88
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>89
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>89
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>90
<211>65
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>90
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcu65
<210>91
<211>65
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>91
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcu65
<210>92
<211>65
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>92
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcu65
<210>93
<211>65
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>93
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcu65
<210>94
<211>65
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>94
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcu65
<210>95
<211>65
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>95
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcu65
<210>96
<211>65
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>96
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcu65
<210>97
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>97
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>98
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>98
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>99
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>99
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>100
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>100
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>101
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>101
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>102
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>102
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>103
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>103
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>104
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>104
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>105
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>105
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>106
<211>62
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>106
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gu62
<210>107
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>107
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>108
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>108
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>109
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>109
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>110
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>110
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>111
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>111
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>112
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>112
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>113
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>113
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>114
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>114
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>115
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>115
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>116
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>116
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>117
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>117
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>118
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>118
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>119
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>119
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>120
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>120
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>121
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>121
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>122
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>122
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>123
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>123
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>124
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>124
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>125
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>125
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>126
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>126
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>127
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>127
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>128
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>128
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>129
<211>67
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<223>descriptionofcombineddna/rnamolecule:synthetic
oligonucleotide
<400>129
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcutt67
<210>130
<211>68
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<223>descriptionofcombineddna/rnamolecule:synthetic
oligonucleotide
<400>130
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuut68
<210>131
<211>62
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>131
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gu62
<210>132
<211>63
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<223>descriptionofcombineddna/rnamolecule:synthetic
oligonucleotide
<400>132
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gut63
<210>133
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>133
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>134
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>134
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>135
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>135
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>136
<211>62
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>136
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gu62
<210>137
<211>62
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>137
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gu62
<210>138
<211>67
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<223>descriptionofcombineddna/rnamolecule:synthetic
oligonucleotide
<400>138
agcauagcaagttaaaataaggctagtccgttaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>139
<211>67
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<223>descriptionofcombineddna/rnamolecule:synthetic
oligonucleotide
<400>139
agcauagcaagttaaaataaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>140
<211>67
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<223>descriptionofcombineddna/rnamolecule:synthetic
oligonucleotide
<400>140
agcauagcaaguuaaaauaaggctagtccgttaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>141
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>141
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>142
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>142
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>143
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>143
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>144
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>144
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>145
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>145
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>146
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>146
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>147
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>147
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>148
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>148
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>149
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>149
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>150
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>150
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>151
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>151
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>152
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>152
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>153
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>153
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>154
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>154
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>155
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>155
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>156
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>156
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>157
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>157
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>158
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>158
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>159
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>159
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>160
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>160
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>161
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>161
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>162
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>162
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>163
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>163
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>164
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>164
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>165
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>165
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>166
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>166
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>167
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>167
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>168
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>168
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>169
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>169
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>170
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>170
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>171
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>171
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>172
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>172
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>173
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>173
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>174
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>174
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>175
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>175
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>176
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>176
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>177
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>177
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>178
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>178
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>179
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>179
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>180
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>180
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>181
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>181
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>182
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>182
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>183
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>183
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>184
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>184
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>185
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>185
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>186
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>186
uuauauccaacacuucgugguuuuagagcuaugcu35
<210>187
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>187
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>188
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>188
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>189
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>189
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>190
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>190
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>191
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>191
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>192
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>192
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>193
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>193
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>194
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>194
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>195
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>195
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>196
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>196
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>197
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>197
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>198
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>198
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>199
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>199
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>200
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>200
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>201
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>201
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>202
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>202
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>203
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>203
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>204
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>204
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>205
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>205
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>206
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>206
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>207
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>207
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>208
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>208
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>209
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>209
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>210
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>210
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>211
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>211
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>212
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>212
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>213
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>213
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>214
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>214
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>215
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>215
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>216
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>216
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>217
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>217
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>218
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>218
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>219
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>219
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>220
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>220
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>221
<211>36
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<223>descriptionofcombineddna/rnamolecule:synthetic
oligonucleotide
<400>221
ctuauauccaacacuucgugguuuuagagcuaugct36
<210>222
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>222
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>223
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>223
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>224
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>224
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>225
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>225
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>226
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>226
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>227
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>227
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>228
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>228
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>229
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>229
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>230
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>230
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>231
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>231
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>232
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>232
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>233
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>233
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>234
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>234
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>235
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>235
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>236
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>236
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>237
<211>37
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<223>descriptionofcombineddna/rnamolecule:synthetic
oligonucleotide
<400>237
cuuauauccaacacuucgugguuuuagagcuaugcut37
<210>238
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>238
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>239
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>239
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>240
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>240
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>241
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>241
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>242
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>242
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>243
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>243
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>244
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>244
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>245
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>245
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>246
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>246
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>247
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>247
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>248
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>248
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>249
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>249
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>250
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>250
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>251
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>251
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>252
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>252
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>253
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>253
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>254
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>254
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>255
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>255
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>256
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>256
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>257
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>257
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>258
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>258
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>259
<211>35
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>259
cuuauauccaacacuucgugguuuuagagcuaugc35
<210>260
<211>34
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>260
cuuauauccaacacuucgugguuuuagagcuaug34
<210>261
<211>33
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>261
cuuauauccaacacuucgugguuuuagagcuau33
<210>262
<211>32
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>262
cuuauauccaacacuucgugguuuuagagcua32
<210>263
<211>31
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>263
cuuauauccaacacuucgugguuuuagagcu31
<210>264
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>264
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>265
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>265
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>266
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>266
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>267
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>267
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>268
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>268
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>269
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>269
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>270
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>270
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>271
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>271
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>272
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>272
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>273
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>273
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>274
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>274
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>275
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>275
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>276
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>276
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>277
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>277
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>278
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>278
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>279
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>279
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>280
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>280
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>281
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>281
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>282
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>282
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>283
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>283
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>284
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>284
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>285
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>285
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>286
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>286
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>287
<211>33
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>287
auauccaacacuucgugguuuuagagcuaugcu33
<210>288
<211>32
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>288
uauccaacacuucgugguuuuagagcuaugcu32
<210>289
<211>33
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<223>descriptionofcombineddna/rnamolecule:synthetic
oligonucleotide
<400>289
atauccaacacuucgugguuuuagagcuaugcu33
<210>290
<211>32
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<223>descriptionofcombineddna/rnamolecule:synthetic
oligonucleotide
<400>290
tauccaacacuucgugguuuuagagcuaugcu32
<210>291
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>291
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>292
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>292
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>293
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>293
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>294
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>294
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>295
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>295
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>296
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>296
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>297
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>297
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>298
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>298
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>299
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>299
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>300
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>300
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>301
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>301
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>302
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>302
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>303
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>303
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>304
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>304
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>305
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>305
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>306
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>306
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>307
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>307
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>308
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>308
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>309
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>309
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>310
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>310
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>311
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>311
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>312
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>312
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>313
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>313
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>314
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>314
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>315
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>315
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>316
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>316
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>317
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>317
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>318
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>318
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>319
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>319
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>320
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>320
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>321
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>321
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>322
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>322
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>323
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>323
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>324
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>324
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>325
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>325
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>326
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>326
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>327
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>327
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>328
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>328
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>329
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>329
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>330
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>330
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>331
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>331
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>332
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>332
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>333
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>333
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>334
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>334
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>335
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>335
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>336
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>336
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>337
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>337
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>338
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>338
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>339
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>339
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>340
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>340
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>341
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>341
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>342
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>342
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>343
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>343
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>344
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>344
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>345
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>345
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>346
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>346
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>347
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>347
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>348
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>348
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>349
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>349
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>350
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>350
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>351
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>351
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>352
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>352
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>353
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>353
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>354
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>354
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>355
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>355
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>356
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>356
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>357
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>357
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>358
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>358
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>359
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>359
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>360
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>360
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>361
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>361
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>362
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>362
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>363
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>363
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>364
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>364
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>365
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>365
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>366
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>366
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>367
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>367
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>368
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>368
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>369
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>369
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>370
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>370
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>371
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>371
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>372
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>372
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>373
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>373
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>374
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>374
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>375
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>375
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>376
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>376
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>377
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>377
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>378
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>378
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>379
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>379
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>380
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>380
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>381
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>381
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>382
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>382
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>383
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>383
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>384
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>384
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>385
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>385
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>386
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>386
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>387
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>387
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>388
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>388
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>389
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>389
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>390
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>390
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>391
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>391
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>392
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>392
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>393
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>393
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>394
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>394
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>395
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>395
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>396
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>396
uccauuucauagucuuuccuguuuuagagcuaugcu36
<210>397
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>397
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>398
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>398
cuuagagaauauuuguagagguuuuagagcuaugcu36
<210>399
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>399
uugacuauaaugaauacuucguuuuagagcuaugcu36
<210>400
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>400
caaaacacgcauaaaaauuuguuuuagagcuaugcu36
<210>401
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>401
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>402
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>402
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>403
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>403
ggcuuauauccaacacuucgguuuuagagcuaugcu36
<210>404
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>404
auuucacauaaaacucuuuuguuuuagagcuaugcu36
<210>405
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>405
ucaaauuaugaggugcuggaguuuuagagcuaugcu36
<210>406
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>406
uacagcuuuaugugacuaauguuuuagagcuaugcu36
<210>407
<211>1367
<212>prt
<213>streptococcuspyogenes
<400>407
metasplyslystyrserileglyleuaspileglythrasnserval
151015
glytrpalavalilethraspglutyrlysvalproserlyslysphe
202530
lysvalleuglyasnthrasparghisserilelyslysasnleuile
354045
glyalaleuleupheaspserglygluthralaglualathrargleu
505560
lysargthralaargargargtyrthrargarglysasnargilecys
65707580
tyrleuglngluilepheserasnglumetalalysvalaspaspser
859095
phephehisargleuglugluserpheleuvalglugluasplyslys
100105110
hisgluarghisproilepheglyasnilevalaspgluvalalatyr
115120125
hisglulystyrprothriletyrhisleuarglyslysleuvalasp
130135140
serthrasplysalaaspleuargleuiletyrleualaleualahis
145150155160
metilelyspheargglyhispheleuilegluglyaspleuasnpro
165170175
aspasnseraspvalasplysleupheileglnleuvalglnthrtyr
180185190
asnglnleupheglugluasnproileasnalaserglyvalaspala
195200205
lysalaileleuseralaargleuserlysserargargleugluasn
210215220
leuilealaglnleuproglyglulyslysasnglyleupheglyasn
225230235240
leuilealaleuserleuglyleuthrproasnphelysserasnphe
245250255
aspleualagluaspalalysleuglnleuserlysaspthrtyrasp
260265270
aspaspleuaspasnleuleualaglnileglyaspglntyralaasp
275280285
leupheleualaalalysasnleuseraspalaileleuleuserasp
290295300
ileleuargvalasnthrgluilethrlysalaproleuseralaser
305310315320
metilelysargtyraspgluhishisglnaspleuthrleuleulys
325330335
alaleuvalargglnglnleuproglulystyrlysgluilephephe
340345350
aspglnserlysasnglytyralaglytyrileaspglyglyalaser
355360365
glnglugluphetyrlyspheilelysproileleuglulysmetasp
370375380
glythrglugluleuleuvallysleuasnarggluaspleuleuarg
385390395400
lysglnargthrpheaspasnglyserileprohisglnilehisleu
405410415
glygluleuhisileleuargargglngluaspphetyrpropheleu
420425430
lysaspasnargglulysileglulysileleuthrpheargilepro
435440445
tyrtyrvalglyproleualaargglyasnserargphealatrpmet
450455460
thrarglysserglugluthrilethrprotrpasnpheglugluval
465470475480
valasplysglyalaseralaglnserpheilegluargmetthrasn
485490495
pheasplysasnleuproasnglulysvalleuprolyshisserleu
500505510
leutyrglutyrphethrvaltyrasngluleuthrlysvallystyr
515520525
valthrgluglymetarglysproalapheleuserglygluglnlys
530535540
lysalailevalaspleuleuphelysthrasnarglysvalthrval
545550555560
lysglnleulysgluasptyrphelyslysileglucyspheaspser
565570575
valgluileserglyvalgluaspargpheasnalaserleuglythr
580585590
tyrhisaspleuleulysileilelysasplysasppheleuaspasn
595600605
glugluasngluaspileleugluaspilevalleuthrleuthrleu
610615620
phegluaspargglumetileglugluargleulysthrtyralahis
625630635640
leupheaspasplysvalmetlysglnleulysargargargtyrthr
645650655
glytrpglyargleuserarglysleuileasnglyileargasplys
660665670
glnserglylysthrileleuasppheleulysseraspglypheala
675680685
asnargasnphemetglnleuilehisaspaspserleuthrphelys
690695700
gluaspileglnlysalaglnvalserglyglnglyaspserleuhis
705710715720
gluhisilealaasnleualaglyserproalailelyslysglyile
725730735
leuglnthrvallysvalvalaspgluleuvallysvalmetglyarg
740745750
hislysprogluasnilevalileglumetalaarggluasnglnthr
755760765
thrglnlysglyglnlysasnserarggluargmetlysargileglu
770775780
gluglyilelysgluleuglyserglnileleulysgluhisproval
785790795800
gluasnthrglnleuglnasnglulysleutyrleutyrtyrleugln
805810815
asnglyargaspmettyrvalaspglngluleuaspileasnargleu
820825830
serasptyraspvalasphisilevalproglnserpheleulysasp
835840845
aspserileaspasnlysvalleuthrargserasplysasnarggly
850855860
lysseraspasnvalproserglugluvalvallyslysmetlysasn
865870875880
tyrtrpargglnleuleuasnalalysleuilethrglnarglysphe
885890895
aspasnleuthrlysalagluargglyglyleusergluleuasplys
900905910
alaglypheilelysargglnleuvalgluthrargglnilethrlys
915920925
hisvalalaglnileleuaspserargmetasnthrlystyraspglu
930935940
asnasplysleuilearggluvallysvalilethrleulysserlys
945950955960
leuvalseraspphearglysasppheglnphetyrlysvalargglu
965970975
ileasnasntyrhishisalahisaspalatyrleuasnalavalval
980985990
glythralaleuilelyslystyrprolysleugluserglupheval
99510001005
tyrglyasptyrlysvaltyraspvalarglysmetilealalys
101010151020
sergluglngluileglylysalathralalystyrphephetyr
102510301035
serasnilemetasnphephelysthrgluilethrleualaasn
104010451050
glygluilearglysargproleuilegluthrasnglygluthr
105510601065
glygluilevaltrpasplysglyargaspphealathrvalarg
107010751080
lysvalleusermetproglnvalasnilevallyslysthrglu
108510901095
valglnthrglyglypheserlysgluserileleuprolysarg
110011051110
asnserasplysleuilealaarglyslysasptrpaspprolys
111511201125
lystyrglyglypheaspserprothrvalalatyrservalleu
113011351140
valvalalalysvalglulysglylysserlyslysleulysser
114511501155
vallysgluleuleuglyilethrilemetgluargserserphe
116011651170
glulysasnproileasppheleuglualalysglytyrlysglu
117511801185
vallyslysaspleuileilelysleuprolystyrserleuphe
119011951200
gluleugluasnglyarglysargmetleualaseralaglyglu
120512101215
leuglnlysglyasngluleualaleuproserlystyrvalasn
122012251230
pheleutyrleualaserhistyrglulysleulysglyserpro
123512401245
gluaspasngluglnlysglnleuphevalgluglnhislyshis
125012551260
tyrleuaspgluileilegluglnileserglupheserlysarg
126512701275
valileleualaaspalaasnleuasplysvalleuseralatyr
128012851290
asnlyshisargasplysproilearggluglnalagluasnile
129513001305
ilehisleuphethrleuthrasnleuglyalaproalaalaphe
131013151320
lystyrpheaspthrthrileasparglysargtyrthrserthr
132513301335
lysgluvalleuaspalathrleuilehisglnserilethrgly
134013451350
leutyrgluthrargileaspleuserglnleuglyglyasp
135513601365
<210>408
<211>1416
<212>prt
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
polypeptide
<400>408
metglyserseralaprolyslyslysarglysvalglyilehisgly
151015
valproalaalametasplyslystyrserileglyleuaspilegly
202530
thrasnservalglytrpalavalilethraspglutyrlysvalpro
354045
serlyslysphelysvalleuglyasnthrasparghisserilelys
505560
lysasnleuileglyalaleuleupheaspserglygluthralaglu
65707580
alathrargleulysargthralaargargargtyrthrargarglys
859095
asnargilecystyrleuglngluilepheserasnglumetalalys
100105110
valaspaspserphephehisargleuglugluserpheleuvalglu
115120125
gluasplyslyshisgluarghisproilepheglyasnilevalasp
130135140
gluvalalatyrhisglulystyrprothriletyrhisleuarglys
145150155160
lysleuvalaspserthrasplysalaaspleuargleuiletyrleu
165170175
alaleualahismetilelyspheargglyhispheleuileglugly
180185190
aspleuasnproaspasnseraspvalasplysleupheileglnleu
195200205
valglnthrtyrasnglnleupheglugluasnproileasnalaser
210215220
glyvalaspalalysalaileleuseralaargleuserlysserarg
225230235240
argleugluasnleuilealaglnleuproglyglulyslysasngly
245250255
leupheglyasnleuilealaleuserleuglyleuthrproasnphe
260265270
lysserasnpheaspleualagluaspalalysleuglnleuserlys
275280285
aspthrtyraspaspaspleuaspasnleuleualaglnileglyasp
290295300
glntyralaaspleupheleualaalalysasnleuseraspalaile
305310315320
leuleuseraspileleuargvalasnthrgluilethrlysalapro
325330335
leuseralasermetilelysargtyraspgluhishisglnaspleu
340345350
thrleuleulysalaleuvalargglnglnleuproglulystyrlys
355360365
gluilephepheaspglnserlysasnglytyralaglytyrileasp
370375380
glyglyalaserglnglugluphetyrlyspheilelysproileleu
385390395400
glulysmetaspglythrglugluleuleuvallysleuasnargglu
405410415
aspleuleuarglysglnargthrpheaspasnglyserileprohis
420425430
glnilehisleuglygluleuhisalaileleuargargglngluasp
435440445
phetyrpropheleulysaspasnargglulysileglulysileleu
450455460
thrpheargileprotyrtyrvalglyproleualaargglyasnser
465470475480
argphealatrpmetthrarglysserglugluthrilethrprotrp
485490495
asnpheglugluvalvalasplysglyalaseralaglnserpheile
500505510
gluargmetthrasnpheasplysasnleuproasnglulysvalleu
515520525
prolyshisserleuleutyrglutyrphethrvaltyrasngluleu
530535540
thrlysvallystyrvalthrgluglymetarglysproalapheleu
545550555560
serglygluglnlyslysalailevalaspleuleuphelysthrasn
565570575
arglysvalthrvallysglnleulysgluasptyrphelyslysile
580585590
glucyspheaspservalgluileserglyvalgluaspargpheasn
595600605
alaserleuglythrtyrhisaspleuleulysileilelysasplys
610615620
asppheleuaspasnglugluasngluaspileleugluaspileval
625630635640
leuthrleuthrleuphegluaspargglumetileglugluargleu
645650655
lysthrtyralahisleupheaspasplysvalmetlysglnleulys
660665670
argargargtyrthrglytrpglyargleuserarglysleuileasn
675680685
glyileargasplysglnserglylysthrileleuasppheleulys
690695700
seraspglyphealaasnargasnphemetglnleuilehisaspasp
705710715720
serleuthrphelysgluaspileglnlysalaglnvalserglygln
725730735
glyaspserleuhisgluhisilealaasnleualaglyserproala
740745750
ilelyslysglyileleuglnthrvallysvalvalaspgluleuval
755760765
lysvalmetglyarghislysprogluasnilevalileglumetala
770775780
arggluasnglnthrthrglnlysglyglnlysasnserarggluarg
785790795800
metlysargileglugluglyilelysgluleuglyserglnileleu
805810815
lysgluhisprovalgluasnthrglnleuglnasnglulysleutyr
820825830
leutyrtyrleuglnasnglyargaspmettyrvalaspglngluleu
835840845
aspileasnargleuserasptyraspvalasphisilevalprogln
850855860
serpheleulysaspaspserileaspasnlysvalleuthrargser
865870875880
asplysasnargglylysseraspasnvalproserglugluvalval
885890895
lyslysmetlysasntyrtrpargglnleuleuasnalalysleuile
900905910
thrglnarglyspheaspasnleuthrlysalagluargglyglyleu
915920925
sergluleuasplysalaglypheilelysargglnleuvalgluthr
930935940
argglnilethrlyshisvalalaglnileleuaspserargmetasn
945950955960
thrlystyraspgluasnasplysleuilearggluvallysvalile
965970975
thrleulysserlysleuvalseraspphearglysasppheglnphe
980985990
tyrlysvalarggluileasnasntyrhishisalahisaspalatyr
99510001005
leuasnalavalvalglythralaleuilelyslystyrprolys
101010151020
leuglusergluphevaltyrglyasptyrlysvaltyraspval
102510301035
arglysmetilealalyssergluglngluileglylysalathr
104010451050
alalystyrphephetyrserasnilemetasnphephelysthr
105510601065
gluilethrleualaasnglygluilearglysargproleuile
107010751080
gluthrasnglygluthrglygluilevaltrpasplysglyarg
108510901095
aspphealathrvalarglysvalleusermetproglnvalasn
110011051110
ilevallyslysthrgluvalglnthrglyglypheserlysglu
111511201125
serileleuprolysargasnserasplysleuilealaarglys
113011351140
lysasptrpaspprolyslystyrglyglypheaspserprothr
114511501155
valalatyrservalleuvalvalalalysvalglulysglylys
116011651170
serlyslysleulysservallysgluleuleuglyilethrile
117511801185
metgluargserserpheglulysasnproileasppheleuglu
119011951200
alalysglytyrlysgluvallyslysaspleuileilelysleu
120512101215
prolystyrserleuphegluleugluasnglyarglysargmet
122012251230
leualaseralaglygluleuglnlysglyasngluleualaleu
123512401245
proserlystyrvalasnpheleutyrleualaserhistyrglu
125012551260
lysleulysglyserprogluaspasngluglnlysglnleuphe
126512701275
valgluglnhislyshistyrleuaspgluileilegluglnile
128012851290
serglupheserlysargvalileleualaaspalaasnleuasp
129513001305
lysvalleuseralatyrasnlyshisargasplysproilearg
131013151320
gluglnalagluasnileilehisleuphethrleuthrasnleu
132513301335
glyalaproalaalaphelystyrpheaspthrthrileasparg
134013451350
lysargtyrthrserthrlysgluvalleuaspalathrleuile
135513601365
hisglnserilethrglyleutyrgluthrargileaspleuser
137013751380
glnleuglyglyaspalaalaprolyslyslysarglysvalasp
138513901395
prolyslyslysarglysvalalaalaalaleugluhishishis
140014051410
hishishis
1415
<210>409
<211>4251
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
polynucleotide
<400>409
atgggcagcagcgccccaaagaagaagcggaaggtcggtatccacggagtcccagcagcc60
atggacaaaaagtactctattggcctggatatcgggaccaacagcgtcgggtgggctgtt120
atcaccgacgagtataaagtaccttcgaaaaagttcaaagtgctgggcaacaccgatcgc180
cattcaatcaaaaagaacttgattggtgcgctgttgtttgactccggggaaaccgccgag240
gcgactcgccttaaacgtacagcacgtcgccggtacactcggcgtaagaatcgcatttgc300
tatttgcaggaaatctttagcaacgagatggcaaaagtcgatgactcgtttttccaccgc360
ctcgaggaaagctttctggtggaggaagacaaaaagcatgagcgtcacccgatcttcggc420
aacattgtcgatgaagtagcgtatcatgaaaaatacccaaccatttaccacttacgcaaa480
aagctggtggacagcactgacaaagctgatttgcgccttatctatttagccctggcacat540
atgattaagtttcgtggtcacttcctgatcgaaggagacttaaatcccgacaacagtgat600
gttgataaattgtttattcagcttgtccaaacttacaatcaactgttcgaggaaaacccg660
atcaatgcctccggtgtggatgcaaaagccattttaagtgcacgccttagcaagtcccgt720
cgcttagaaaaccttatcgcgcagctgcccggcgagaaaaagaatggtttgtttgggaac780
cttattgccttgagcttaggcctcaccccgaatttcaaaagtaatttcgatcttgcagaa840
gacgccaaattacaactgtcgaaggatacttatgatgacgatctcgataatctgttagcg900
cagattggtgaccaatacgccgatctttttctggcggctaaaaatctgagcgacgccatc960
ttgctttcggatattctccgcgttaacaccgaaatcacgaaagcgcctcttagtgccagc1020
atgattaaacgttatgatgaacaccaccaggacctgaccttactcaaagcgttggttcgc1080
cagcaactgccagagaagtacaaagaaatcttctttgatcagtcaaagaatggttatgcc1140
ggctatattgacgggggtgcaagccaagaggaattctacaaatttatcaagcctattctg1200
gagaaaatggatggcaccgaagagttattggtgaagcttaaccgtgaagacctcctgcgg1260
aaacagcgcacattcgataatggttcgatcccacaccaaatccatttgggggagttacac1320
gctattttgcgtcgccaggaagacttttaccctttcctgaaggataaccgggagaaaatt1380
gagaagatccttacctttcgtattccgtattacgtaggccccttagcacggggtaatagc1440
cgtttcgcgtggatgacacggaagtcggaagagacgatcaccccgtggaacttcgaagag1500
gtagtcgacaagggcgcatcagcgcagtcttttattgaacgtatgacgaatttcgataaa1560
aacttgcccaatgagaaggtgcttccgaaacattccttgttatatgaatattttacagtt1620
tacaacgagctgaccaaggttaaatacgtgacggaaggaatgcgcaagcccgcttttctt1680
agcggtgagcaaaaaaaggcgatcgtcgacctgttattcaaaacgaatcgtaaggtgact1740
gtaaagcaactcaaagaagattacttcaaaaagattgagtgcttcgacagcgtcgaaatc1800
tctggggtagaggatcggtttaacgcaagtttaggtacctaccatgacctgcttaaaatc1860
attaaggataaagacttcttagataatgaagagaacgaagatattctcgaggacatcgtc1920
ttgacgttaaccttatttgaggatcgtgaaatgattgaggaacgcctcaaaacttatgcc1980
cacctgttcgacgataaggtgatgaagcagctgaaacgtcggcgctacacaggatggggc2040
cgcttgagtcgcaaacttattaacggaatccgtgacaagcaatccggcaaaacgattctg2100
gatttcttgaagtcggacggatttgctaatcgcaacttcatgcagttgatccatgatgac2160
tccctgacttttaaagaggatattcaaaaggcgcaggttagtggtcaaggcgacagctta2220
cacgaacacatcgcaaatttggctggttcgccggccattaaaaaggggatcctccagacc2280
gtgaaagttgtagatgagcttgttaaggtcatgggtcgtcataagcccgaaaacatcgtg2340
attgaaatggcgcgggagaatcaaacgacccagaaaggacaaaagaatagccgtgaacgg2400
atgaagcggatcgaggaaggcattaaagagctggggtctcaaatcttgaaggaacaccct2460
gtggagaacactcagctccaaaatgaaaaactttacctgtactatttgcagaacggacgc2520
gatatgtacgtggaccaagagttggatattaatcggctgagtgactacgacgttgatcat2580
atcgtcccgcagagcttcctcaaagacgattctattgacaataaggtactgacgcgctct2640
gataaaaaccgtggtaagtcggacaacgtgccctccgaagaggttgtgaaaaagatgaaa2700
aattattggcgccagcttttaaacgcgaagctgatcacacaacgtaaattcgataatttg2760
accaaggctgaacggggtggcctgagcgagttagataaggcaggatttattaaacgccag2820
ttagtggagactcgtcaaatcaccaaacatgtcgcgcagattttggacagccggatgaac2880
accaagtacgatgaaaatgacaaactgatccgtgaggtgaaagtcattactctgaagtcc2940
aaattagttagtgatttccggaaggactttcaattctacaaagtccgtgaaattaataac3000
tatcatcacgcacatgacgcgtacctgaatgcagtggttgggaccgcccttatcaagaaa3060
tatcctaagctggagtcggagtttgtctatggcgactataaggtatacgatgttcgcaaa3120
atgattgcgaaatctgagcaggagatcggtaaggcaaccgcaaaatatttcttttactca3180
aacattatgaatttctttaagacagaaatcactctggccaacggggagattcgcaaacgt3240
ccgttgatcgaaacaaacggcgagactggcgaaattgtttgggacaaagggcgtgatttc3300
gcgacggtgcgcaaggtactgagcatgcctcaagtcaatattgttaagaaaaccgaagtg3360
cagacgggcgggttttccaaggaaagcatcttacccaaacgtaattcagataaacttatt3420
gcacgcaaaaaggactgggatccgaaaaagtatggaggcttcgacagtccaaccgtagcc3480
tactctgttctcgttgtagcgaaagtagaaaagggtaaatccaagaaactgaaatctgtc3540
aaggagttgcttggaatcaccattatggagcgtagctccttcgagaagaacccgattgac3600
tttctggaagccaaaggatataaagaggtcaagaaagatcttatcattaagctgcctaag3660
tattcactcttcgagctggaaaatggtcgtaaacgcatgctcgcttctgccggcgagttg3720
cagaagggcaatgaattagcacttccatcaaagtacgttaacttcctgtatttggccagc3780
cattacgagaaactgaaggggtctccagaggacaacgaacagaaacaattatttgtagag3840
cagcacaagcattatcttgatgaaatcattgagcaaatttccgaattcagtaaacgcgta3900
atcctggccgatgcaaacctcgacaaggtgctgagcgcttacaataagcatcgcgacaaa3960
cctatccgtgagcaggctgaaaatatcattcacctgttcacattaacgaacctgggcgct4020
ccggccgcttttaaatatttcgacacgacaatcgaccgtaagcgctataccagtacgaaa4080
gaagtgttggatgcgacccttattcaccagtcaattacaggattatatgagacccgtatc4140
gaccttagccaattaggtggggatgcggccccgaagaaaaaacgcaaagtggatccgaag4200
aaaaaacgcaaagtggcggccgcactcgagcaccaccaccaccaccactga4251
<210>410
<211>1429
<212>prt
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
polypeptide
<400>410
metglylysproileproasnproleuleuglyleuaspserthrala
151015
prolyslyslysarglysvalglyilehisglyvalproalaalaasp
202530
lyslystyrserileglyleuaspileglythrasnservalglytrp
354045
alavalilethraspglutyrlysvalproserlyslysphelysval
505560
leuglyasnthrasparghisserilelyslysasnleuileglyala
65707580
leuleupheaspserglygluthralaglualathrargleulysarg
859095
thralaargargargtyrthrargarglysasnargilecystyrleu
100105110
glngluilepheserasnglumetalalysvalaspaspserphephe
115120125
hisargleuglugluserpheleuvalglugluasplyslyshisglu
130135140
arghisproilepheglyasnilevalaspgluvalalatyrhisglu
145150155160
lystyrprothriletyrhisleuarglyslysleuvalaspserthr
165170175
asplysalaaspleuargleuiletyrleualaleualahismetile
180185190
lyspheargglyhispheleuilegluglyaspleuasnproaspasn
195200205
seraspvalasplysleupheileglnleuvalglnthrtyrasngln
210215220
leupheglugluasnproileasnalaserglyvalaspalalysala
225230235240
ileleuseralaargleuserlysserargargleugluasnleuile
245250255
alaglnleuproglyglulyslysasnglyleupheglyasnleuile
260265270
alaleuserleuglyleuthrproasnphelysserasnpheaspleu
275280285
alagluaspalalysleuglnleuserlysaspthrtyraspaspasp
290295300
leuaspasnleuleualaglnileglyaspglntyralaaspleuphe
305310315320
leualaalalysasnleuseraspalaileleuleuseraspileleu
325330335
argvalasnthrgluilethrlysalaproleuseralasermetile
340345350
lysargtyraspgluhishisglnaspleuthrleuleulysalaleu
355360365
valargglnglnleuproglulystyrlysgluilephepheaspgln
370375380
serlysasnglytyralaglytyrileaspglyglyalaserglnglu
385390395400
gluphetyrlyspheilelysproileleuglulysmetaspglythr
405410415
glugluleuleuvallysleuasnarggluaspleuleuarglysgln
420425430
argthrpheaspasnglyserileprohisglnilehisleuglyglu
435440445
leuhisalaileleuargargglngluaspphetyrpropheleulys
450455460
aspasnargglulysileglulysileleuthrpheargileprotyr
465470475480
tyrvalglyproleualaargglyasnserargphealatrpmetthr
485490495
arglysserglugluthrilethrprotrpasnpheglugluvalval
500505510
asplysglyalaseralaglnserpheilegluargmetthrasnphe
515520525
asplysasnleuproasnglulysvalleuprolyshisserleuleu
530535540
tyrglutyrphethrvaltyrasngluleuthrlysvallystyrval
545550555560
thrgluglymetarglysproalapheleuserglygluglnlyslys
565570575
alailevalaspleuleuphelysthrasnarglysvalthrvallys
580585590
glnleulysgluasptyrphelyslysileglucyspheaspserval
595600605
gluileserglyvalgluaspargpheasnalaserleuglythrtyr
610615620
hisaspleuleulysileilelysasplysasppheleuaspasnglu
625630635640
gluasngluaspileleugluaspilevalleuthrleuthrleuphe
645650655
gluaspargglumetileglugluargleulysthrtyralahisleu
660665670
pheaspasplysvalmetlysglnleulysargargargtyrthrgly
675680685
trpglyargleuserarglysleuileasnglyileargasplysgln
690695700
serglylysthrileleuasppheleulysseraspglyphealaasn
705710715720
argasnphemetglnleuilehisaspaspserleuthrphelysglu
725730735
aspileglnlysalaglnvalserglyglnglyaspserleuhisglu
740745750
hisilealaasnleualaglyserproalailelyslysglyileleu
755760765
glnthrvallysvalvalaspgluleuvallysvalmetglyarghis
770775780
lysprogluasnilevalileglumetalaarggluasnglnthrthr
785790795800
glnlysglyglnlysasnserarggluargmetlysargilegluglu
805810815
glyilelysgluleuglyserglnileleulysgluhisprovalglu
820825830
asnthrglnleuglnasnglulysleutyrleutyrtyrleuglnasn
835840845
glyargaspmettyrvalaspglngluleuaspileasnargleuser
850855860
asptyraspvalasphisilevalproglnserpheleulysaspasp
865870875880
serileaspasnlysvalleuthrargserasplysasnargglylys
885890895
seraspasnvalproserglugluvalvallyslysmetlysasntyr
900905910
trpargglnleuleuasnalalysleuilethrglnarglyspheasp
915920925
asnleuthrlysalagluargglyglyleusergluleuasplysala
930935940
glypheilelysargglnleuvalgluthrargglnilethrlyshis
945950955960
valalaglnileleuaspserargmetasnthrlystyraspgluasn
965970975
asplysleuilearggluvallysvalilethrleulysserlysleu
980985990
valseraspphearglysasppheglnphetyrlysvalarggluile
99510001005
asnasntyrhishisalahisaspalatyrleuasnalavalval
101010151020
glythralaleuilelyslystyrprolysleuglusergluphe
102510301035
valtyrglyasptyrlysvaltyraspvalarglysmetileala
104010451050
lyssergluglngluileglylysalathralalystyrphephe
105510601065
tyrserasnilemetasnphephelysthrgluilethrleuala
107010751080
asnglygluilearglysargproleuilegluthrasnglyglu
108510901095
thrglygluilevaltrpasplysglyargaspphealathrval
110011051110
arglysvalleusermetproglnvalasnilevallyslysthr
111511201125
gluvalglnthrglyglypheserlysgluserileleuprolys
113011351140
argasnserasplysleuilealaarglyslysasptrpasppro
114511501155
lyslystyrglyglypheaspserprothrvalalatyrserval
116011651170
leuvalvalalalysvalglulysglylysserlyslysleulys
117511801185
servallysgluleuleuglyilethrilemetgluargserser
119011951200
pheglulysasnproileasppheleuglualalysglytyrlys
120512101215
gluvallyslysaspleuileilelysleuprolystyrserleu
122012251230
phegluleugluasnglyarglysargmetleualaseralagly
123512401245
gluleuglnlysglyasngluleualaleuproserlystyrval
125012551260
asnpheleutyrleualaserhistyrglulysleulysglyser
126512701275
progluaspasngluglnlysglnleuphevalgluglnhislys
128012851290
histyrleuaspgluileilegluglnileserglupheserlys
129513001305
argvalileleualaaspalaasnleuasplysvalleuserala
131013151320
tyrasnlyshisargasplysproilearggluglnalagluasn
132513301335
ileilehisleuphethrleuthrasnleuglyalaproalaala
134013451350
phelystyrpheaspthrthrileasparglysargtyrthrser
135513601365
thrlysgluvalleuaspalathrleuilehisglnserilethr
137013751380
glyleutyrgluthrargileaspleuserglnleuglyglyasp
138513901395
serargalaaspprolyslyslysarglysvalgluphehishis
140014051410
thrglyleuvalaspproserservalproserleuserleuasn
141514201425
arg
<210>411
<211>4290
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
polynucleotide
<400>411
atgggcaagcccatccctaaccccctgttggggctggacagcaccgctcccaaaaagaaa60
aggaaggtgggcattcacggcgtgcctgcggccgacaaaaagtacagcatcggccttgat120
atcggcaccaatagcgtgggctgggccgttatcacagacgaatacaaggtacccagcaag180
aagttcaaggtgctggggaatacagacaggcactctatcaagaaaaaccttatcggggct240
ctgctgtttgactcaggcgagaccgccgaggccaccaggttgaagaggaccgcaaggcga300
aggtacacccggaggaagaacaggatctgctatctgcaggagatcttcagcaacgagatg360
gccaaggtggacgacagcttcttccacaggctggaggagagcttccttgtcgaggaggat420
aagaagcacgaacgacaccccatcttcggcaacatagtcgacgaggtcgcttatcacgag480
aagtaccccaccatctaccacctgcgaaagaaattggtggatagcaccgataaagccgac540
ttgcgacttatctacttggctctggcgcacatgattaagttcaggggccacttcctgatc600
gagggcgaccttaaccccgacaacagtgacgtagacaaattgttcatccagcttgtacag660
acctataaccagctgttcgaggaaaaccctattaacgccagcggggtggatgcgaaggcc720
atacttagcgccaggctgagcaaaagcaggcgcttggagaacctgatagcccagctgccc780
ggtgaaaagaagaacggcctcttcggtaatctgattgccctgagcctgggcctgaccccc840
aacttcaagagcaacttcgacctggcagaagatgccaagctgcagttgagtaaggacacc900
tatgacgacgacttggacaatctgctcgcccaaatcggcgaccagtacgctgacctgttc960
ctcgccgccaagaacctttctgacgcaatcctgcttagcgatatccttagggtgaacaca1020
gagatcaccaaggcccccctgagcgccagcatgatcaagaggtacgacgagcaccatcag1080
gacctgacccttctgaaggccctggtgaggcagcaactgcccgagaagtacaaggagatc1140
tttttcgaccagagcaagaacggctacgccggctacatcgacggcggagccagccaagag1200
gagttctacaagttcatcaagcccatcctggagaagatggatggcaccgaggagctgctg1260
gtgaagctgaacagggaagatttgctccggaagcagaggacctttgacaacggtagcatc1320
ccccaccagatccacctgggcgagctgcacgcaatactgaggcgacaggaggatttctac1380
cccttcctcaaggacaatagggagaaaatcgaaaagattctgaccttcaggatcccctac1440
tacgtgggccctcttgccaggggcaacagccgattcgcttggatgacaagaaagagcgag1500
gagaccatcaccccctggaacttcgaggaagtggtggacaaaggagcaagcgcgcagtct1560
ttcatcgaacggatgaccaatttcgacaaaaacctgcctaacgagaaggtgctgcccaag1620
cacagcctgctttacgagtacttcaccgtgtacaacgagctcaccaaggtgaaatatgtg1680
accgagggcatgcgaaaacccgctttcctgagcggcgagcagaagaaggccatcgtggac1740
ctgctgttcaagaccaacaggaaggtgaccgtgaagcagctgaaggaggactacttcaag1800
aagatcgagtgctttgatagcgtggaaataagcggcgtggaggacaggttcaacgccagc1860
ctgggcacctaccacgacttgttgaagataatcaaagacaaggatttcctggataatgag1920
gagaacgaggatatactcgaggacatcgtgctgactttgaccctgtttgaggaccgagag1980
atgattgaagaaaggctcaaaacctacgcccacctgttcgacgacaaagtgatgaaacaa2040
ctgaagagacgaagatacaccggctggggcagactgtccaggaagctcatcaacggcatt2100
agggacaagcagagcggcaagaccatcctggatttcctgaagtccgacggcttcgccaac2160
cgaaacttcatgcagctgattcacgatgacagcttgaccttcaaggaggacatccagaag2220
gcccaggttagcggccagggcgactccctgcacgaacatattgcaaacctggcaggctcc2280
cctgcgatcaagaagggcatactgcagaccgttaaggttgtggacgaattggtcaaggtc2340
atgggcaggcacaagcccgaaaacatagttatagagatggccagagagaaccagaccacc2400
caaaagggccagaagaacagccgggagcgcatgaaaaggatcgaggagggtatcaaggaa2460
ctcggaagccagatcctcaaagagcaccccgtggagaatacccagctccagaacgagaag2520
ctgtacctgtactacctgcagaacggcagggacatgtacgttgaccaggagttggacatc2580
aacaggctttcagactatgacgtggatcacatagtgccccagagctttcttaaagacgat2640
agcatcgacaacaaggtcctgacccgctccgacaaaaacaggggcaaaagcgacaacgtg2700
ccaagcgaagaggtggttaaaaagatgaagaactactggaggcaactgctcaacgcgaaa2760
ttgatcacccagagaaagttcgataacctgaccaaggccgagaggggcggactctccgaa2820
cttgacaaagcgggcttcataaagaggcagctggtcgagacccgacagatcacgaagcac2880
gtggcccaaatcctcgacagcagaatgaataccaagtacgatgagaatgacaaactcatc2940
agggaagtgaaagtgattaccctgaagagcaagttggtgtccgactttcgcaaagatttc3000
cagttctacaaggtgagggagatcaacaactaccaccatgcccacgacgcatacctgaac3060
gccgtggtcggcaccgccctgattaagaagtatccaaagctggagtccgaatttgtctac3120
ggcgactacaaagtttacgatgtgaggaagatgatcgctaagagcgaacaggagatcggc3180
aaggccaccgctaagtatttcttctacagcaacatcatgaactttttcaagaccgagatc3240
acacttgccaacggcgaaatcaggaagaggccgcttatcgagaccaacggtgagaccggc3300
gagatcgtgtgggacaagggcagggacttcgccaccgtgaggaaagtcctgagcatgccc3360
caggtgaatattgtgaaaaaaactgaggtgcagacaggcggctttagcaaggaatccatc3420
ctgcccaagaggaacagcgacaagctgatcgcccggaagaaggactgggaccctaagaag3480
tatggaggcttcgacagccccaccgtagcctacagcgtgctggtggtcgcgaaggtagag3540
aaggggaagagcaagaaactgaagagcgtgaaggagctgctcggcataaccatcatggag3600
aggtccagctttgagaagaaccccattgactttttggaagccaagggctacaaagaggtc3660
aaaaaggacctgatcatcaaactccccaagtactccctgtttgaattggagaacggcaga3720
aagaggatgctggcgagcgctggggaactgcaaaagggcaacgaactggcgctgcccagc3780
aagtacgtgaattttctgtacctggcgtcccactacgaaaagctgaaaggcagccccgag3840
gacaacgagcagaagcagctgttcgtggagcagcacaagcattacctggacgagataatc3900
gagcaaatcagcgagttcagcaagagggtgattctggccgacgcgaacctggataaggtc3960
ctcagcgcctacaacaagcaccgagacaaacccatcagggagcaggccgagaatatcata4020
cacctgttcaccctgacaaatctgggcgcacctgcggcattcaaatacttcgataccacc4080
atcgacaggaaaaggtacactagcactaaggaggtgctggatgccaccttgatccaccag4140
tccattaccggcctgtatgagaccaggatcgacctgagccagcttggaggcgactctagg4200
gcggacccaaaaaagaaaaggaaggtggaattccaccacactggactagtggatccgagc4260
tcggtaccaagcttaagtttaaaccgctga4290
<210>412
<211>4601
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
polynucleotide
<400>412
taatacgactcactatagggagacccaagctggctagcgtttaaacgggccctctagact60
cgagcggccgccaccatgggcaagcccatccctaaccccctgttggggctggacagcacc120
gctcccaaaaagaaaaggaaggtgggcattcacggcgtgcctgcggccgacaaaaagtac180
agcatcggccttgatatcggcaccaatagcgtgggctgggccgttatcacagacgaatac240
aaggtacccagcaagaagttcaaggtgctggggaatacagacaggcactctatcaagaaa300
aaccttatcggggctctgctgtttgactcaggcgagaccgccgaggccaccaggttgaag360
aggaccgcaaggcgaaggtacacccggaggaagaacaggatctgctatctgcaggagatc420
ttcagcaacgagatggccaaggtggacgacagcttcttccacaggctggaggagagcttc480
cttgtcgaggaggataagaagcacgaacgacaccccatcttcggcaacatagtcgacgag540
gtcgcttatcacgagaagtaccccaccatctaccacctgcgaaagaaattggtggatagc600
accgataaagccgacttgcgacttatctacttggctctggcgcacatgattaagttcagg660
ggccacttcctgatcgagggcgaccttaaccccgacaacagtgacgtagacaaattgttc720
atccagcttgtacagacctataaccagctgttcgaggaaaaccctattaacgccagcggg780
gtggatgcgaaggccatacttagcgccaggctgagcaaaagcaggcgcttggagaacctg840
atagcccagctgcccggtgaaaagaagaacggcctcttcggtaatctgattgccctgagc900
ctgggcctgacccccaacttcaagagcaacttcgacctggcagaagatgccaagctgcag960
ttgagtaaggacacctatgacgacgacttggacaatctgctcgcccaaatcggcgaccag1020
tacgctgacctgttcctcgccgccaagaacctttctgacgcaatcctgcttagcgatatc1080
cttagggtgaacacagagatcaccaaggcccccctgagcgccagcatgatcaagaggtac1140
gacgagcaccatcaggacctgacccttctgaaggccctggtgaggcagcaactgcccgag1200
aagtacaaggagatctttttcgaccagagcaagaacggctacgccggctacatcgacggc1260
ggagccagccaagaggagttctacaagttcatcaagcccatcctggagaagatggatggc1320
accgaggagctgctggtgaagctgaacagggaagatttgctccggaagcagaggaccttt1380
gacaacggtagcatcccccaccagatccacctgggcgagctgcacgcaatactgaggcga1440
caggaggatttctaccccttcctcaaggacaatagggagaaaatcgaaaagattctgacc1500
ttcaggatcccctactacgtgggccctcttgccaggggcaacagccgattcgcttggatg1560
acaagaaagagcgaggagaccatcaccccctggaacttcgaggaagtggtggacaaagga1620
gcaagcgcgcagtctttcatcgaacggatgaccaatttcgacaaaaacctgcctaacgag1680
aaggtgctgcccaagcacagcctgctttacgagtacttcaccgtgtacaacgagctcacc1740
aaggtgaaatatgtgaccgagggcatgcgaaaacccgctttcctgagcggcgagcagaag1800
aaggccatcgtggacctgctgttcaagaccaacaggaaggtgaccgtgaagcagctgaag1860
gaggactacttcaagaagatcgagtgctttgatagcgtggaaataagcggcgtggaggac1920
aggttcaacgccagcctgggcacctaccacgacttgttgaagataatcaaagacaaggat1980
ttcctggataatgaggagaacgaggatatactcgaggacatcgtgctgactttgaccctg2040
tttgaggaccgagagatgattgaagaaaggctcaaaacctacgcccacctgttcgacgac2100
aaagtgatgaaacaactgaagagacgaagatacaccggctggggcagactgtccaggaag2160
ctcatcaacggcattagggacaagcagagcggcaagaccatcctggatttcctgaagtcc2220
gacggcttcgccaaccgaaacttcatgcagctgattcacgatgacagcttgaccttcaag2280
gaggacatccagaaggcccaggttagcggccagggcgactccctgcacgaacatattgca2340
aacctggcaggctcccctgcgatcaagaagggcatactgcagaccgttaaggttgtggac2400
gaattggtcaaggtcatgggcaggcacaagcccgaaaacatagttatagagatggccaga2460
gagaaccagaccacccaaaagggccagaagaacagccgggagcgcatgaaaaggatcgag2520
gagggtatcaaggaactcggaagccagatcctcaaagagcaccccgtggagaatacccag2580
ctccagaacgagaagctgtacctgtactacctgcagaacggcagggacatgtacgttgac2640
caggagttggacatcaacaggctttcagactatgacgtggatcacatagtgccccagagc2700
tttcttaaagacgatagcatcgacaacaaggtcctgacccgctccgacaaaaacaggggc2760
aaaagcgacaacgtgccaagcgaagaggtggttaaaaagatgaagaactactggaggcaa2820
ctgctcaacgcgaaattgatcacccagagaaagttcgataacctgaccaaggccgagagg2880
ggcggactctccgaacttgacaaagcgggcttcataaagaggcagctggtcgagacccga2940
cagatcacgaagcacgtggcccaaatcctcgacagcagaatgaataccaagtacgatgag3000
aatgacaaactcatcagggaagtgaaagtgattaccctgaagagcaagttggtgtccgac3060
tttcgcaaagatttccagttctacaaggtgagggagatcaacaactaccaccatgcccac3120
gacgcatacctgaacgccgtggtcggcaccgccctgattaagaagtatccaaagctggag3180
tccgaatttgtctacggcgactacaaagtttacgatgtgaggaagatgatcgctaagagc3240
gaacaggagatcggcaaggccaccgctaagtatttcttctacagcaacatcatgaacttt3300
ttcaagaccgagatcacacttgccaacggcgaaatcaggaagaggccgcttatcgagacc3360
aacggtgagaccggcgagatcgtgtgggacaagggcagggacttcgccaccgtgaggaaa3420
gtcctgagcatgccccaggtgaatattgtgaaaaaaactgaggtgcagacaggcggcttt3480
agcaaggaatccatcctgcccaagaggaacagcgacaagctgatcgcccggaagaaggac3540
tgggaccctaagaagtatggaggcttcgacagccccaccgtagcctacagcgtgctggtg3600
gtcgcgaaggtagagaaggggaagagcaagaaactgaagagcgtgaaggagctgctcggc3660
ataaccatcatggagaggtccagctttgagaagaaccccattgactttttggaagccaag3720
ggctacaaagaggtcaaaaaggacctgatcatcaaactccccaagtactccctgtttgaa3780
ttggagaacggcagaaagaggatgctggcgagcgctggggaactgcaaaagggcaacgaa3840
ctggcgctgcccagcaagtacgtgaattttctgtacctggcgtcccactacgaaaagctg3900
aaaggcagccccgaggacaacgagcagaagcagctgttcgtggagcagcacaagcattac3960
ctggacgagataatcgagcaaatcagcgagttcagcaagagggtgattctggccgacgcg4020
aacctggataaggtcctcagcgcctacaacaagcaccgagacaaacccatcagggagcag4080
gccgagaatatcatacacctgttcaccctgacaaatctgggcgcacctgcggcattcaaa4140
tacttcgataccaccatcgacaggaaaaggtacactagcactaaggaggtgctggatgcc4200
accttgatccaccagtccattaccggcctgtatgagaccaggatcgacctgagccagctt4260
ggaggcgactctagggcggacccaaaaaagaaaaggaaggtggaattccaccacactgga4320
ctagtggatccgagctcggtaccaagcttaagtttaaaccgctgatcagcctcgactgtg4380
ccttctagttgccagccatctgttgtttgcccctcccccgtgccttccttgaccctggaa4440
ggtgccactcccactgtcctttcctaataaaatgaggaaattgcatcgcattgtctgagt4500
aggtgtcattctattctggggggtggggtggggcaggacagcaagggggaggattgggaa4560
gacaatagcaggcatgctggggatgcggtgggctctatggc4601
<210>413
<211>4584
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
polynucleotide
<400>413
gggagacccaagcuggcuagcguuuaaacgggcccucuagacucgagcggccgccaccau60
gggcaagcccaucccuaacccccuguuggggcuggacagcaccgcucccaaaaagaaaag120
gaaggugggcauucacggcgugccugcggccgacaaaaaguacagcaucggccuugauau180
cggcaccaauagcgugggcugggccguuaucacagacgaauacaagguacccagcaagaa240
guucaaggugcuggggaauacagacaggcacucuaucaagaaaaaccuuaucggggcucu300
gcuguuugacucaggcgagaccgccgaggccaccagguugaagaggaccgcaaggcgaag360
guacacccggaggaagaacaggaucugcuaucugcaggagaucuucagcaacgagauggc420
caagguggacgacagcuucuuccacaggcuggaggagagcuuccuugucgaggaggauaa480
gaagcacgaacgacaccccaucuucggcaacauagucgacgaggucgcuuaucacgagaa540
guaccccaccaucuaccaccugcgaaagaaauugguggauagcaccgauaaagccgacuu600
gcgacuuaucuacuuggcucuggcgcacaugauuaaguucaggggccacuuccugaucga660
gggcgaccuuaaccccgacaacagugacguagacaaauuguucauccagcuuguacagac720
cuauaaccagcuguucgaggaaaacccuauuaacgccagcgggguggaugcgaaggccau780
acuuagcgccaggcugagcaaaagcaggcgcuuggagaaccugauagcccagcugcccgg840
ugaaaagaagaacggccucuucgguaaucugauugcccugagccugggccugacccccaa900
cuucaagagcaacuucgaccuggcagaagaugccaagcugcaguugaguaaggacaccua960
ugacgacgacuuggacaaucugcucgcccaaaucggcgaccaguacgcugaccuguuccu1020
cgccgccaagaaccuuucugacgcaauccugcuuagcgauauccuuagggugaacacaga1080
gaucaccaaggccccccugagcgccagcaugaucaagagguacgacgagcaccaucagga1140
ccugacccuucugaaggcccuggugaggcagcaacugcccgagaaguacaaggagaucuu1200
uuucgaccagagcaagaacggcuacgccggcuacaucgacggcggagccagccaagagga1260
guucuacaaguucaucaagcccauccuggagaagauggauggcaccgaggagcugcuggu1320
gaagcugaacagggaagauuugcuccggaagcagaggaccuuugacaacgguagcauccc1380
ccaccagauccaccugggcgagcugcacgcaauacugaggcgacaggaggauuucuaccc1440
cuuccucaaggacaauagggagaaaaucgaaaagauucugaccuucaggauccccuacua1500
cgugggcccucuugccaggggcaacagccgauucgcuuggaugacaagaaagagcgagga1560
gaccaucacccccuggaacuucgaggaagugguggacaaaggagcaagcgcgcagucuuu1620
caucgaacggaugaccaauuucgacaaaaaccugccuaacgagaaggugcugcccaagca1680
cagccugcuuuacgaguacuucaccguguacaacgagcucaccaaggugaaauaugugac1740
cgagggcaugcgaaaacccgcuuuccugagcggcgagcagaagaaggccaucguggaccu1800
gcuguucaagaccaacaggaaggugaccgugaagcagcugaaggaggacuacuucaagaa1860
gaucgagugcuuugauagcguggaaauaagcggcguggaggacagguucaacgccagccu1920
gggcaccuaccacgacuuguugaagauaaucaaagacaaggauuuccuggauaaugagga1980
gaacgaggauauacucgaggacaucgugcugacuuugacccuguuugaggaccgagagau2040
gauugaagaaaggcucaaaaccuacgcccaccuguucgacgacaaagugaugaaacaacu2100
gaagagacgaagauacaccggcuggggcagacuguccaggaagcucaucaacggcauuag2160
ggacaagcagagcggcaagaccauccuggauuuccugaaguccgacggcuucgccaaccg2220
aaacuucaugcagcugauucacgaugacagcuugaccuucaaggaggacauccagaaggc2280
ccagguuagcggccagggcgacucccugcacgaacauauugcaaaccuggcaggcucccc2340
ugcgaucaagaagggcauacugcagaccguuaagguuguggacgaauuggucaaggucau2400
gggcaggcacaagcccgaaaacauaguuauagagauggccagagagaaccagaccaccca2460
aaagggccagaagaacagccgggagcgcaugaaaaggaucgaggaggguaucaaggaacu2520
cggaagccagauccucaaagagcaccccguggagaauacccagcuccagaacgagaagcu2580
guaccuguacuaccugcagaacggcagggacauguacguugaccaggaguuggacaucaa2640
caggcuuucagacuaugacguggaucacauagugccccagagcuuucuuaaagacgauag2700
caucgacaacaagguccugacccgcuccgacaaaaacaggggcaaaagcgacaacgugcc2760
aagcgaagaggugguuaaaaagaugaagaacuacuggaggcaacugcucaacgcgaaauu2820
gaucacccagagaaaguucgauaaccugaccaaggccgagaggggcggacucuccgaacu2880
ugacaaagcgggcuucauaaagaggcagcuggucgagacccgacagaucacgaagcacgu2940
ggcccaaauccucgacagcagaaugaauaccaaguacgaugagaaugacaaacucaucag3000
ggaagugaaagugauuacccugaagagcaaguugguguccgacuuucgcaaagauuucca3060
guucuacaaggugagggagaucaacaacuaccaccaugcccacgacgcauaccugaacgc3120
cguggucggcaccgcccugauuaagaaguauccaaagcuggaguccgaauuugucuacgg3180
cgacuacaaaguuuacgaugugaggaagaugaucgcuaagagcgaacaggagaucggcaa3240
ggccaccgcuaaguauuucuucuacagcaacaucaugaacuuuuucaagaccgagaucac3300
acuugccaacggcgaaaucaggaagaggccgcuuaucgagaccaacggugagaccggcga3360
gaucgugugggacaagggcagggacuucgccaccgugaggaaaguccugagcaugcccca3420
ggugaauauugugaaaaaaacugaggugcagacaggcggcuuuagcaaggaauccauccu3480
gcccaagaggaacagcgacaagcugaucgcccggaagaaggacugggacccuaagaagua3540
uggaggcuucgacagccccaccguagccuacagcgugcugguggucgcgaagguagagaa3600
ggggaagagcaagaaacugaagagcgugaaggagcugcucggcauaaccaucauggagag3660
guccagcuuugagaagaaccccauugacuuuuuggaagccaagggcuacaaagaggucaa3720
aaaggaccugaucaucaaacuccccaaguacucccuguuugaauuggagaacggcagaaa3780
gaggaugcuggcgagcgcuggggaacugcaaaagggcaacgaacuggcgcugcccagcaa3840
guacgugaauuuucuguaccuggcgucccacuacgaaaagcugaaaggcagccccgagga3900
caacgagcagaagcagcuguucguggagcagcacaagcauuaccuggacgagauaaucga3960
gcaaaucagcgaguucagcaagagggugauucuggccgacgcgaaccuggauaagguccu4020
cagcgccuacaacaagcaccgagacaaacccaucagggagcaggccgagaauaucauaca4080
ccuguucacccugacaaaucugggcgcaccugcggcauucaaauacuucgauaccaccau4140
cgacaggaaaagguacacuagcacuaaggaggugcuggaugccaccuugauccaccaguc4200
cauuaccggccuguaugagaccaggaucgaccugagccagcuuggaggcgacucuagggc4260
ggacccaaaaaagaaaaggaagguggaauuccaccacacuggacuaguggauccgagcuc4320
gguaccaagcuuaaguuuaaaccgcugaucagccucgacugugccuucuaguugccagcc4380
aucuguuguuugccccucccccgugccuuccuugacccuggaaggugccacucccacugu4440
ccuuuccuaauaaaaugaggaaauugcaucgcauugucugaguaggugucauucuauucu4500
ggggggugggguggggcaggacagcaagggggaggauugggaagacaauagcaggcaugc4560
uggggaugcggugggcucuauggc4584
<210>414
<211>104
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
polynucleotide
<400>414
gcuuauauccaacacuucgugguuuuagagcuagaaauagcaaguuaaaauaaggcuagu60
ccguuaucaacuugaaaaaguggcaccgagucggugcuuuuuuu104
<210>415
<211>21
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>415
gactttgctttccttggtcag21
<210>416
<211>24
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>416
ggcttatatccaacacttcgtggg24
<210>417
<211>9
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>417
atggtcaag9
<210>418
<211>18
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>418
tgtgcagaaggatggagt18
<210>419
<211>20
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>419
ctggtgcttctctcaggata20
<210>420
<211>9
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>420
tggaatatg9
<210>421
<211>24
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
primer
<400>421
ctcttcttgaactggtgctgtctg24
<210>422
<211>25
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
primer
<400>422
caggatgaatccaatggtcatgagg25
<210>423
<211>9
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
probe
<400>423
aagtgcctg9
<210>424
<211>21
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
primer
<400>424
ccattgtctggatttaagcgg21
<210>425
<211>21
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
primer
<400>425
gccacaaaaaatcacaagcca21
<210>426
<211>9
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
probe
<400>426
tttctttgc9
<210>427
<211>64
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>427
agcauagcaaguuaaaauaaggcuaguccgucaacuugaaaaaguggcaccgagucggug60
cuuu64
<210>428
<211>99
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>428
cuuauauccaacacuucgugguuuuagagcuagaaauagcaaguuaaaauaaggcuaguc60
cguuaucaacuugaaaaaguggcaccgagucggugcuuu99
<210>429
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>429
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>430
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>430
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>431
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>431
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>432
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>432
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>433
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>433
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>434
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>434
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>435
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>435
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>436
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>436
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>437
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>437
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>438
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>438
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>439
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>439
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>440
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<220>
<221>modified_base
<222>(1)..(20)
<223>a,c,u,g,unknownorother
<400>440
nnnnnnnnnnnnnnnnnnnnguuuuagagcuaugcu36
<210>441
<211>17
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>441
gtcgcaagcttgctggt17
<210>442
<211>16
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>442
ccctgcgtaaactgga16
<210>443
<211>18
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>443
aacatctgggccctgatt18
<210>444
<211>18
<212>dna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>444
ttcccctaaggcaggctg18
<210>445
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>445
aauuauggggauuacuaggaguuuuagagcuaugcu36
<210>446
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>446
uuuuguaauuaacagcuugcguuuuagagcuaugcu36
<210>447
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>447
ggucacuuuuaacacacccaguuuuagagcuaugcu36
<210>448
<211>36
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>448
cuuauauccaacacuucgugguuuuagagcuaugcu36
<210>449
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>449
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>450
<211>67
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>450
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gugcuuu67
<210>451
<211>62
<212>rna
<213>artificialsequence
<220>
<223>descriptionofartificialsequence:synthetic
oligonucleotide
<400>451
agcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucg60
gu62
- 还没有人留言评论。精彩留言会获得点赞!